1) How to create a new oligo
2) How to open an oligo
3) How to export an oligo
4) How to export oligo data
5) How to align an oligo with a sequence
6) How to create a new sequence
7) How to search a sub-sequence into a sequence
1) How to create a new oligo
To create a new oligonucleotide,
1) Click the Oligo / New Oligo menu
2) Type the name of the new oligo in the Name edit box
3) Type the oligonucleotide's sequence in the Seq edit box
AnnHyb will determinate the length, the molecular weight, the molar extinction coefficient, the GC percent and the melting temperature of the new oligonucleotide
Return
2) How to open an oligo
To open an oligonucleotide,
1) Click Oligo / Open Oligo menu
2) Choose the file where you have saved your oligo (the default extention for saved oligo is OLI
Return
3) How to export an oligo
To save only the selected oligo, click Oligo / Save oligo or Oligo / Save oligo as...
Oligo can be exported in 2 formats: FASTA format and OLI format (AnnHyb). The OLI format contains the salt and primer concentrations for the melting temperature determination while the FASTA format contains only the oligo name and sequence
To save all oligonucleotides in one file, click Oligo / Save all oligo
Multi oligo can be exported in 2 formats: FASTA format and OLA format (AnnHyb). The OLA format contains the salt and primer concentrations for the melting temperature determination while the FASTA format contains only the oligo name and sequence
Return
4) How to export oligo data
To export oligonucleotide's data in a plain text file, click Oligo / Export oligo data or Export data for all oligo
Example of exporting oligo data
Return
5) How to align an oligo with a sequence
To align an oligo with a sequence, you must:
create a new oligo.
Select a color for the oligo to search/align.
Click Oligo / Align Oligo with sequences or Oligo / Quicksearch oligo in sequences menu. The "Align" function allow gap opening in oligo or sequence while the "Quicksearch" function not
Check sequences to align in Select sequences window
Click OK to start the alignment
Results are displayed in a window in the Oligo tab

If you activate the "Automatic check best oligo" option (see Options / Parameters - Align / Quicksearch oligo tab), the aligned oligo are automatically checked if the aligned score is greater than the cut-off
The checked oligo are displayed in a window in the sequence tab with informations on alignment
Oligo are also highlighted on the selected sequence

6) How to create a new sequence
To create a new sequence, click Sequence / New sequence menu
Input the sequence in the Edit window
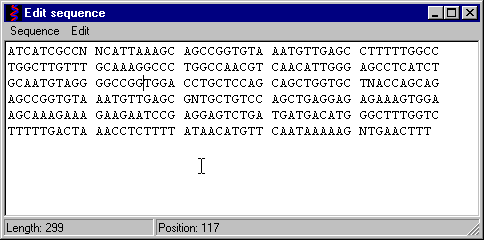
When have typed all nucleotides, click the Sequence / Done menu
Type a sequence name (mandatory) and additional information about the sequence
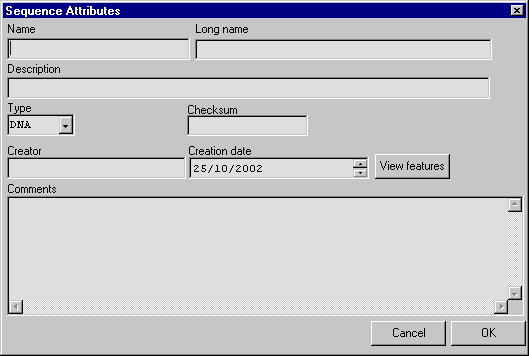
The new sequence is added to the left tree view
Return
6) How to search a sub-sequence into a sequence
Select the sequence to search into, select the Sequence/Quick search for sub-sequence menu option
Type the sub-sequence you want to search
Press the Search button
Return




