Tutorials
How to manipulate an RNA secondary structure?
You have: an RNA secondary structure.
You want: play with it!!
In this tutorial, you will learn how to:
- select secondary interactions and helices
- select single-strands and junctions
- select tertiary interactions
- select contiguous residues
- select non-contiguous residues
Once a secondary structure loaded (for more details, see the tutorial "how to load data into Assemble2?"), it is displayed in the main panel. A secondary structure is made with helices linked and oriented by single-strands and junctions (apical loops, inner loops, 3-way junctions,...). In Assemble2, the helices can be moved to modify the layout of the secondary structure.

Using the mouse, you can:
- zoom in/out the whole secondary structure with the mouse wheel,
- translate the whole secondary structure by keeping the right-mouse pressed on this panel and by dragging the mouse,
- select parts of the secondary structure,
- edit the secondary structure (add/remove helices and/or tertiary interactions) if it is unlocked. (for more details, see the tutorial "how to edit an RNA secondary structure?")
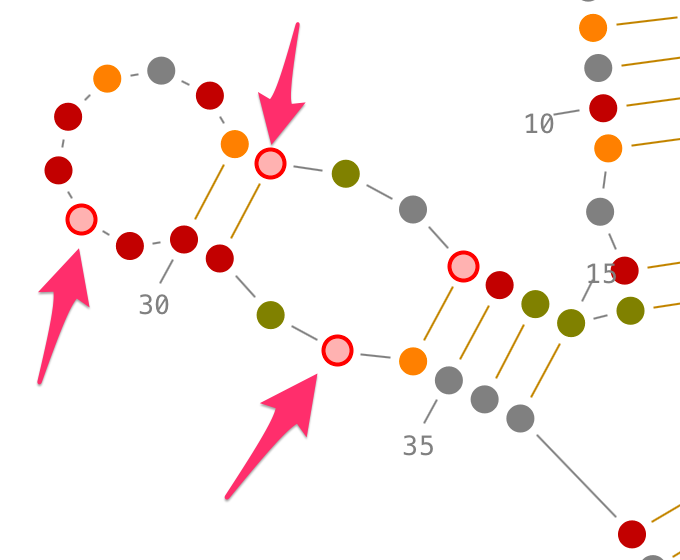
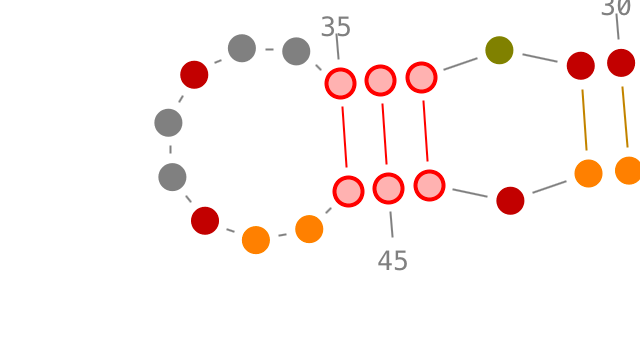
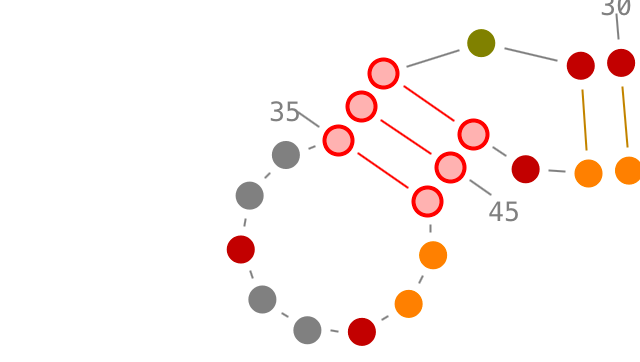
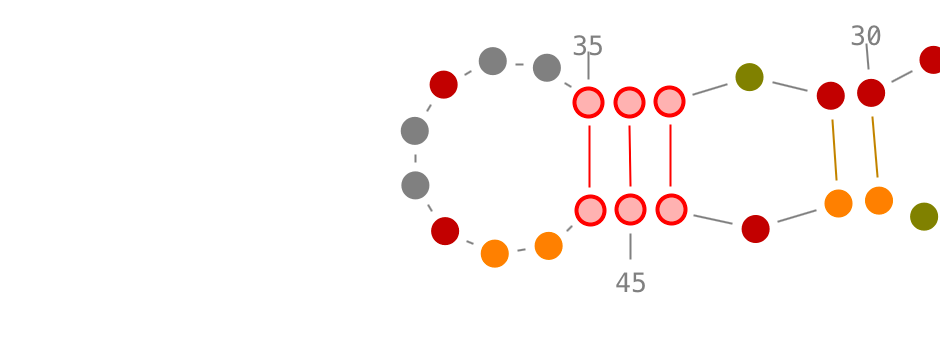
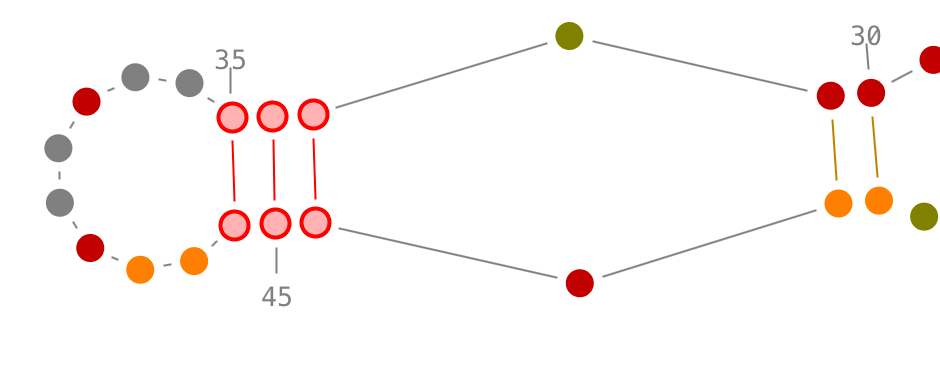
Selection of secondary interactions and helices
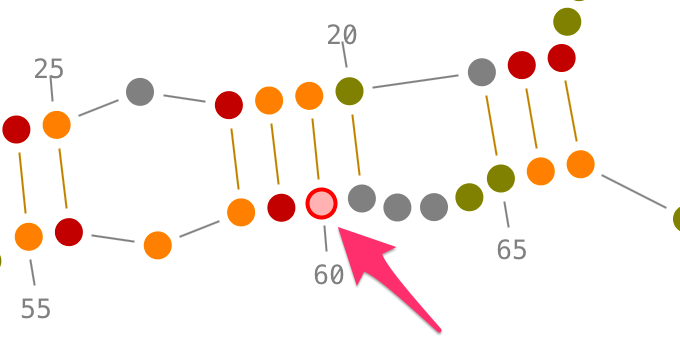
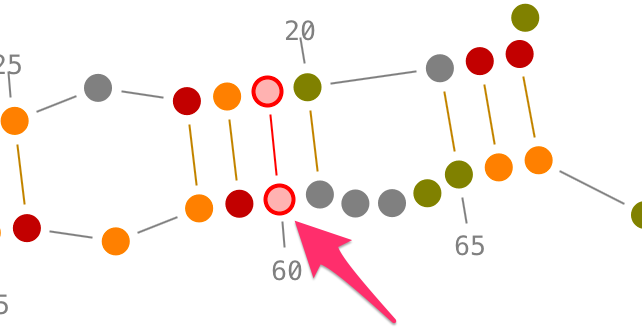
Choose a residue inside the helix you want to select, and left-click on it. By accumulating the mouse clicks on this residue, you will iterate over the selection process:
- one click will select the residue.,
- two consecutive clicks will select its secondary interaction,
- three consecutive clicks will select its helix.
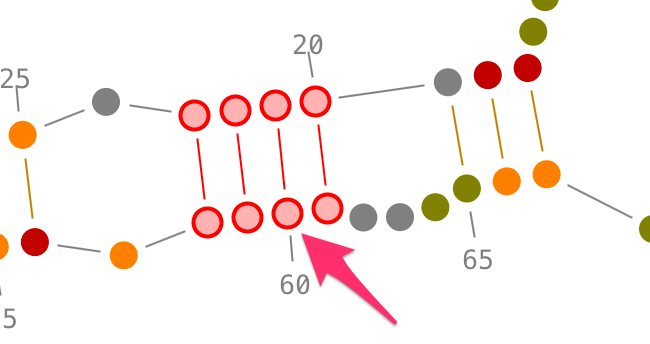
Once an helix selected, you can rotate it:
Selection of single-strands and junctions
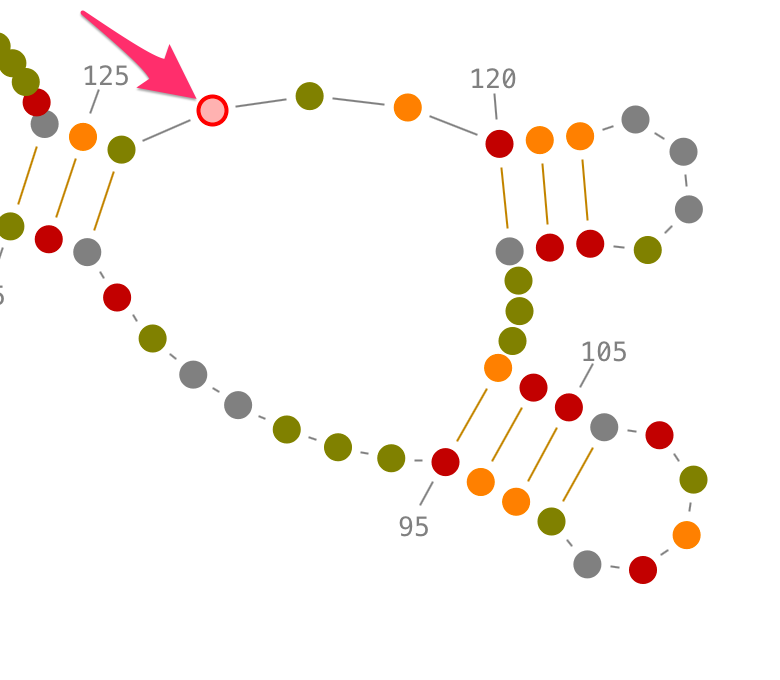
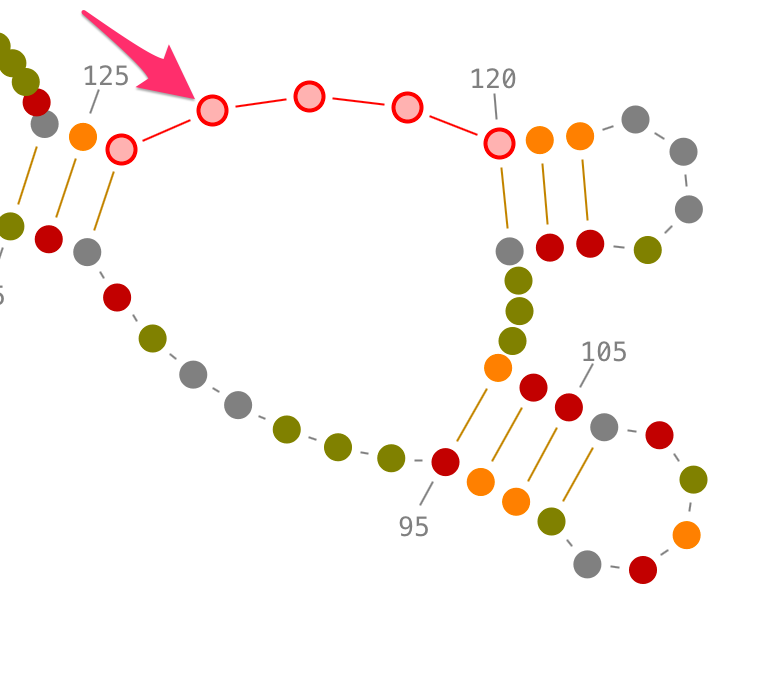
Choose a residue inside the junction or the single-strand you want to select, and left-click on it. By accumulating the mouse clicks on this residue, you will iterate over the selection process:
- one click will select the residue.,
- two consecutive clicks will select its single-strand and pop-up the lateral panel named "3D folds",
- three consecutive clicks will select its junction and pop-up the lateral panel named "3D folds".
Selection of tertiary interactions
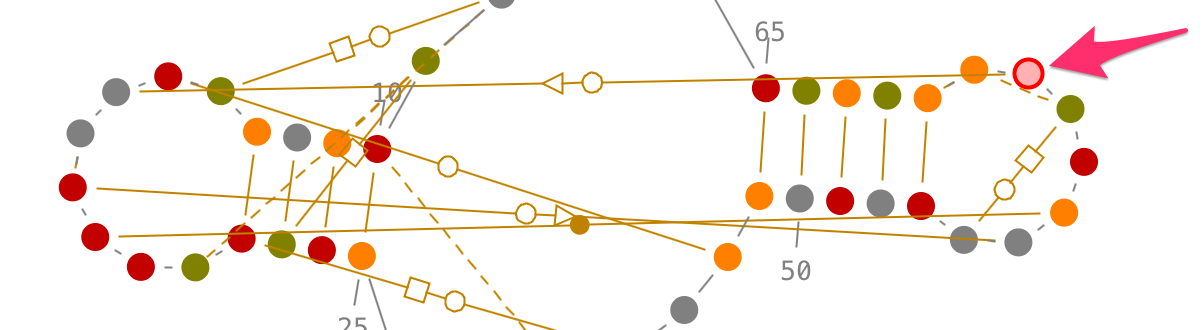
Choose one of the two residues establishing the tertiary interaction, and left-click on it. [Ctrl or Alt] + left-click on the second residue to select the tertiary interaction.
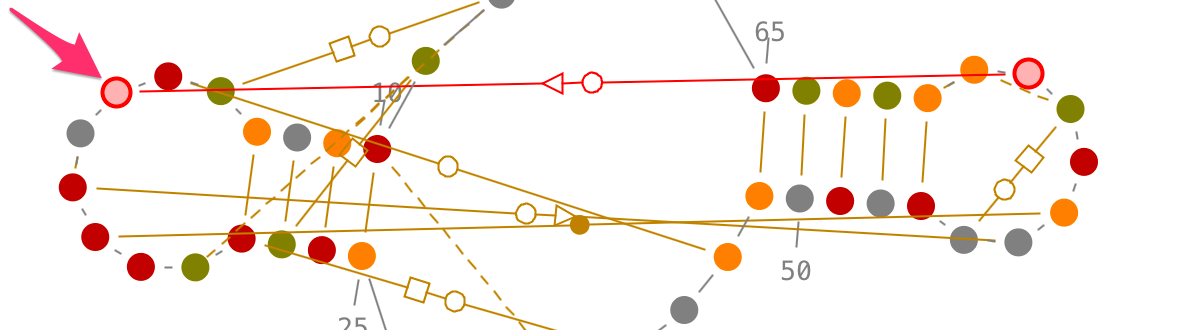
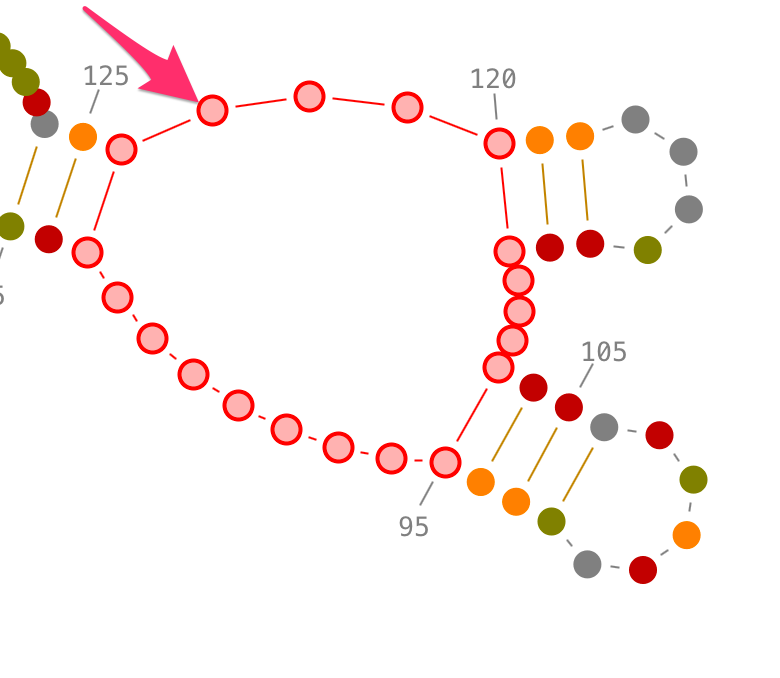
Selection of contiguous residues
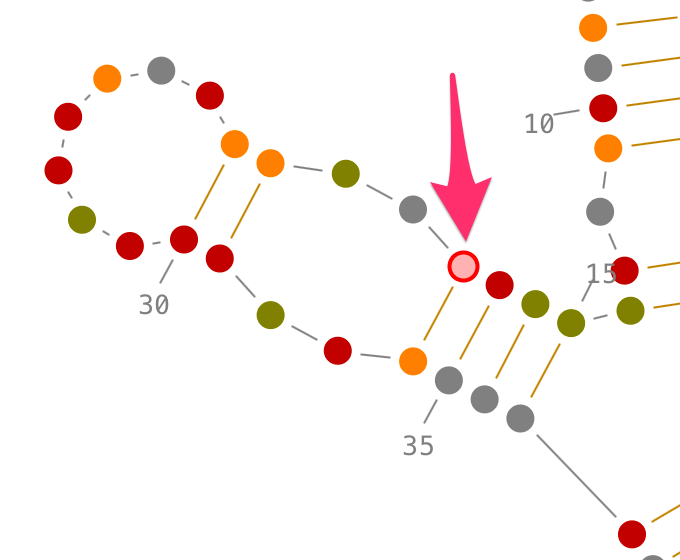
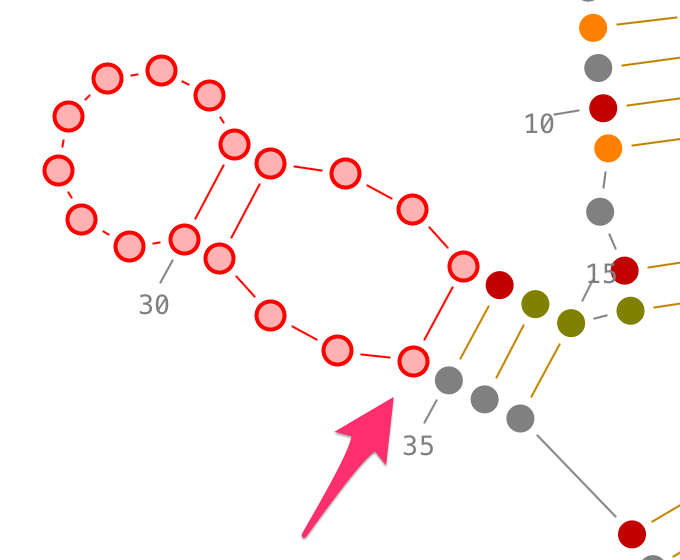
Selection of non-contiguous residues