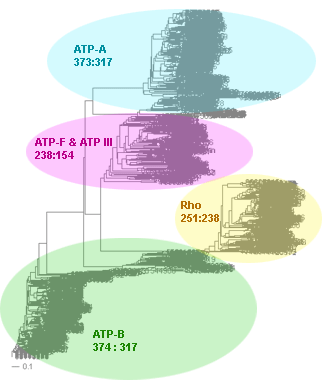
The superfamily consists of ATP synthases subunit A, subunit B, flagellum-specific ATP synthases, transcription termination factor Rho and type III secretion system ATPases.
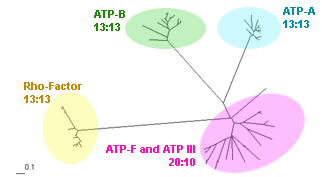
With paramter MANY=8 BrunchClust produces 4 clusters: 3 clusters contain complete (13 members) families of ATP synthases subunit A, subunit B and transcription termination factor Rho, the last one has incomplete cluster with 10 flagellum-specific ATP synthases and type III secretion system ATPases.
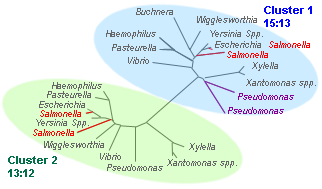
List of 13 gamma proteobacteria: Buchnera aphidicola, Escherichia coli, Haemophilus influenzae, Pasteurella multocida, Pseudomonas aeruginosa, Salmonella typhimurium, Vibrio cholerae, Wigglesworthia glossinidia, Xanthomonas campestris, Xanthomonas axonopodis, Xylella fastidiosa, Yersinia pestis KIM, Yersinia pestis CO92.