Previous: Step 4 - QTL Clustering, Up: Tutorial
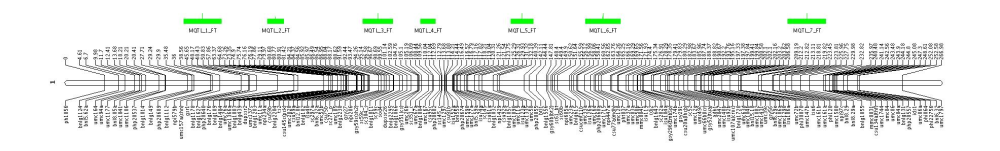
Now, suppose we want to get the 7 meta-QTL of the chromosome 1 separetly on the consensus map. To do so, we can use the command QTLModel as follows:
$java org.metaqtl.main.QTLModel -m meta-map/consensus_map.xml \
-r meta-qtl/meta-analysis_1_res.txt \
-b meta-qtl/meta-analysis_1_best.txt \
-o meta-post/metamap.xml
|
MapView to display the map metamap.xml we obtain something like that:

Finally, it could be useful to project the meta-QTL onto a reference map which provides candidate genes in order to study the congruency between these candidate genes and the meta-QTL.
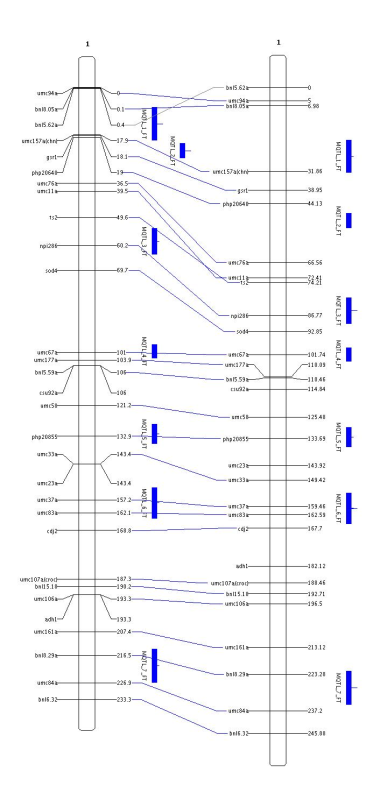
Then, suppose we want to project the meta-QTL onto the reference map genetic_2005 which we have previously used. So, we can use the program QTLProj as follows:
$java org.metaqtl.main.QTLProj -m ref-map/genetic_2005.xml \
-q meta-post/metamap.xml \
-o meta-post/genetic_2005 \
--verbose
|
MMapView to display the result of the projection we obtain something like that: