Next: Step 5 - Post processing, Previous: Step 3 - Projection of the QTL, Up: Tutorial
Now, we are ready to start the QTL meta-analysis itself using the program QTLClust.
$java org.metaqtl.main.QTLClust -q meta-qtl/consensus_map.xml -t xml/ontology.xml \
-k 10 -c 1 -o meta-qtl/meta-analysis_1 --verbose
|
-k to 10 we avoid to explore models with more than 10 meta-QTL on the chromosome.
This command generates three files into the directory meta-qtl:
So, let's have a look to the file meta-analysis_1_model.txt:
Criterion Chromosome Trait Model
AIC 1 FT 7
Criterion Chromosome Trait Model
AICc 1 FT 7
Criterion Chromosome Trait Model
AIC3 1 FT 7
Criterion Chromosome Trait Model
BIC 1 FT 7
Criterion Chromosome Trait Model
AWE 1 FT 7
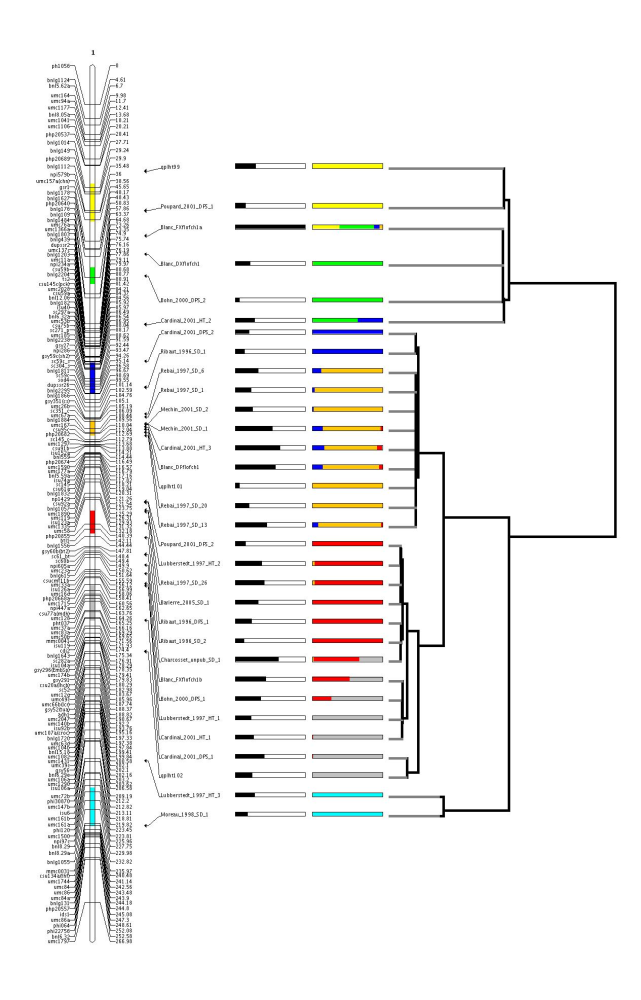
We can see that for each model choice criterion (AIC,AICc,AIC3,BIC,AWE) the best model is the model with 7 QTL clusters, i.e 7 meta-QTL. To see how the observed QTL are distributed into these 7 QTL clusters we can use the program MQTLView. In order to run MQTLView we have to create a file which tells to the program which model it has to represent.
To do so, simply open the file meta-analysis_1_model.txt, keep only one line per chromosome, remove the column called Criterion, and save it as meta-analysis_1_best.txt. Here this file must look like that
Chromosome Trait Model
1 FT 7
Then, we are ready to use MQTLView:
$java org.metaqtl.main.MQTLView -m meta-qtl/consensus_map.xml -c 1 \
-r meta-qtl/meta-analysis_1_res.txt \
-b meta-qtl/meta-analysis_1_best.txt \
-p meta-qtl/images/consensus.par \
-o meta-qtl/images/meta-analysis_1
|

Finally, we can extract for the best model with 7 meta-QTL some useful statistics such like the confidence intervals of the meta-QTL and the a posterori membership probabilities of each observed QTL together with their predicted positions. To do so, use the command QTLClustInfo as follows:
$java org.metaqtl.main.QTLClustInfo -r meta-qtl/meta-analysis_1_res.txt -c 1 \
-o meta-qtl/meta-analysis_1_info.txt \
-b 7 --kmin 1 --kmax 10 -t FT
|
##
# Meta-QTL Table
##
--
QTL Position Weight Distance CI(95%) UCI(95%)
--
1 32.82 0.07 2.21 12.65 12.65
2 57.31 0.1 3.47 5.68 22.48
3 91.52 0.1 1.56 10.07 15.95
4 108.29 0.26 2.74 5.2 5.13
5 139.76 0.24 2.43 7.68 16.89
6 166.86 0.17 7.06 11.75 16.79
7 235.16 0.06 - 12.89 12.89
--
##
# Observed QTL Table
##
--
QTL Position CI(95%) Predicted Memberships 1 --> 7
--
qplht99 22.2 27.75 32.82 1 0 0 0 0 0 0
Poupard_2001_DPS_1 35.48 14.31 32.82 1 0 0 0 0 0 0
Blanc_FXflofch1a 43.91 94 52.53 0.39 0.49 0.08 0.04 0 0 0
Blanc_DXflofch1 50.07 19.99 57.07 0.01 0.99 0 0 0 0 0
Bohn_2000_DPS_2 57.42 6.04 57.31 0 1 0 0 0 0 0
Cardinal_2001_HT_2 73.59 26.11 69.28 0 0.65 0.35 0 0 0 0
Ribaut_1996_SD_1 94.61 12.78 91.52 0 0 1 0 0 0 0
Cardinal_2001_DPS_2 85.91 18.97 91.52 0 0 1 0 0 0 0
Rebai_1997_SD_6 103.51 30.97 106.11 0 0 0.13 0.87 0 0 0
Cardinal_2001_HT_3 107.09 60.09 106.56 0 0 0.17 0.75 0.07 0 0
Mechin_2001_SD_1 106.94 50.06 106.72 0 0 0.15 0.82 0.03 0 0
Blanc_DFflofch1 107.88 54.02 107.35 0 0 0.15 0.8 0.05 0 0
Rebai_1997_SD_1 104.55 21.44 107.79 0 0 0.03 0.97 0 0 0
Mechin_2001_SD_2 106.77 23.83 107.95 0 0 0.02 0.98 0 0 0
Rebai_1997_SD_20 109.73 19.05 108.29 0 0 0 1 0 0 0
qplht101 108.6 5.76 108.29 0 0 0 1 0 0 0
Rebai_1997_SD_13 110.77 42.88 108.98 0 0 0.08 0.9 0.03 0 0
Lubberstedt_1997_HT_2 133.15 35.75 138.82 0 0 0 0.03 0.97 0 0
Barierre_2005_SD_1 136.24 30.97 139.76 0 0 0 0 1 0 0
Poupard_2001_DPS_2 132.94 14.31 139.76 0 0 0 0 1 0 0
Ribaut_1996_SD_2 144.94 12.15 139.76 0 0 0 0 1 0 0
Ribaut_1996_DPS_1 139.97 21.99 139.76 0 0 0 0 1 0 0
Rebai_1997_SD_26 135.75 39.32 140.48 0 0 0 0.03 0.97 0.01 0
Charcosset_unpub_SD_1 150.6 58.33 149.74 0 0 0 0.02 0.65 0.34 0
Blanc_FXflofch1b 154.13 41 152.5 0 0 0 0 0.53 0.47 0
Bohn_2000_DPS_1 157.06 34.69 159.54 0 0 0 0 0.27 0.73 0
Cardinal_2001_DPS_1 172.62 39.16 166.59 0 0 0 0 0.01 0.99 0
Cardinal_2001_HT_1 161.11 25.24 166.59 0 0 0 0 0.01 0.99 0
qplht102 183.06 23.01 166.86 0 0 0 0 0 1 0
Lubberstedt_1997_HT_1 160.36 21.44 166.86 0 0 0 0 0 1 0
Lubberstedt_1997_HT_3 219.66 26.22 235.16 0 0 0 0 0 0 1
Moreau_1998_SD_1 241.43 16.7 235.16 0 0 0 0 0 0 1
--
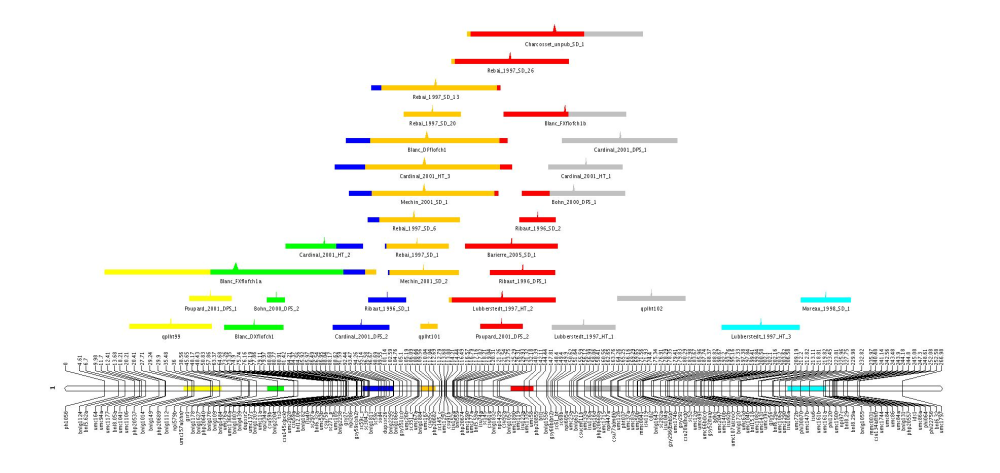
Another techniques to group the observed QTL into clusters is to use standard agglomerative clustering technics, although this method does not make it possible to infer the best number of clusters. Nevertheless, it can help to visualize how the observed QTL are distributed along the chromosome.
Let's run the program QTLTree as follows:
$java org.metaqtl.main.QTLTree -q meta-qtl/consensus_map.xml \
-o meta-qtl/meta-tree.txt \
-t xml/ontology.xml --verbose
|
CR 1
TR FT 32
QT 0 Bohn_2000_DPS_2 57.42 1.54
QT 1 Bohn_2000_DPS_1 157.06 8.85
...
QT 30 Rebai_1997_SD_1 104.55 5.47
QT 31 Mechin_2001_SD_1 106.94 12.77
HC (((((0:0.37,19:0.37):2.37,29:2.37):8.51,8:8.51):51.78,(6:14.63,10:14.63):51.78):1046.37,((((((1:1.02,(7:0.07,15:0.07):1
.02):2.68,26:2.68):48.32,(((2:0,11:0):0.85,(20:0.03,27:0.03):0.85):15.12,(5:2.87,(21:0.64,24:0.64):2.87):15.12):48.32):106
,(4:10.41,9:10.41):106):252.78,((3:8.11,22:8.11):22.26,(((((12:0,18:0):0.04,25:0.04):0.08,28:0.08):0.19,(13:0.01,(16:0,31:
0):0.01):0.19):0.9,(17:0.12,30:0.12):0.9):22.26):252.78):646.19,(14:48.63,23:48.63):646.19):1046.37);
--
At first sight, it is not really easy to imagine from this file the tree that connects the QTL. To display the tree, we can use the command MQTLView. A good point is that we can visualize together the result of the previous clustering and the obtained tree.
$java org.metaqtl.main.MQTLView -m meta-qtl/consensus_map.xml -c 1 \
-r meta-qtl/meta-analysis_1_res.txt \
-b meta-qtl/meta-analysis_1_best.txt \
-p meta-qtl/images/consensus.par \
-o meta-qtl/images/meta-analysis_1
--tree meta-qtl/meta-tree.txt
|