Gff2ps
From Bioinformatics.Org Wiki
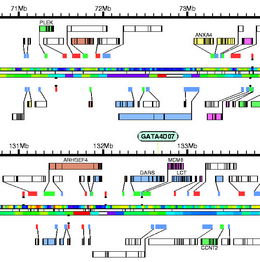
| 
|
Description
Converting genomic annotations in GFF format to PostScript. It scales up from a single gene sequence to múltiple sequences, it can also handle múltiple annotation sources. It was used to produce whole genome maps for different species: Drosophila melanogaster Adams et al. Science 287(5461):2185-2195, 2000, human Venter et al. Science 291(5507):1304-1351, 2001, malaria mosquito Holt et al. Science 298(5591):129-149, 2002, and more recently the map of the diploid genome of a single human individual Levy et al. PLoS Biol 5(10):e254, 2007.
Home page
You can find more information about this tool at:
- http://genome.crg.es/software/gfftools/GFF2PS.html
- http://www1.imim.es/software/gfftools/GFF2PS.html (former URL)
Source code
gff2ps was released under the GNU-GPL software license, and it's available from this link:
- http://genome.imim.es/software/gfftools/GFF2PS.html#DOWNLOADING
- http://www1.imim.es/software/gfftools/GFF2PS.html (former URL)
Executables
It's written as an awk script within a shell script, producing output in PostScript (a programming language for high-quality vector graphics). Depends on bash and GNU-awk.
Supported platforms
Unix/Linux
Documentation
Contact information
Josep F Abril, now at Computational Genomics Lab.