
|
E-CELL2 User's Manual
|
Chapter 5 E-CELL2 Creating User Defined Reactors [ Chapter 1 : Chapter 2 : Chapter 3 : Chapter 4 ][ Top ] |
1 Reactors
E-CELL2 utilizes a combination of three classes of objects to model cell activity: Substances, Reactors, and Systems. Reactors perform the important function of calculating the changes in the amount of Substances over time. Systems express the location of Substances, and Reactors.
Reactors provide the flexibility for users to implement virtually any reaction mechanism in order to meet their needs in modeling specific systems. Reactors are created and compiled separately from the main E-CELL2 application as external object files, and are loaded, as necessary, at runtime. When E-CELL2 loads the Rule Files which specify the simulation model, the necessary Reactors specified in the Rule Files are loaded automatically. Users have the freedom to program their own Reactors in C++ to represent chemical reactions or any other phenomena which may affect the amount of Substances.
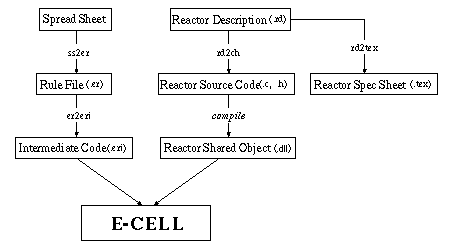
Figure 1. The data flow in the E-CELL2 via file creation and conversion
The above figure shows the data flow in the E-CELL2 system.
As seen in the figure above, Reactors can be created separately from the E-CELL2 core.
To create a Reactor, its specification and actions
must first be written in a Reactor Description file (RD file).
The necessary information is extracted from the RD file to generate
C++ code, which can then be compiled into a dynamic link library which can be coupled with the E-CELL2 on demand(.dll file).
The RD files can also be converted into
Reactor Spec Sheets (in LaTeX format).
1.1 Postern Reactors
There are two types of Reactors in the E-CELL2 , or Normal Reactors and Postern Reactors. Normal Reactors expressed with differential equations are calculated in the E-CELL2 System using integral mechanisms. In contrast, Postern Reactors are calculated independent of integral mechanisms, and are used where differential equations are unsuitable in calculating Substance quantity. This is accomplished through the Multiphase Calculation Structure, where normal reactions are calculated in the React Phase and Postern Reactors in the Posterior Phase.
Postern Reactors may be used under the following circumstances,
- Reactions where Substance quantity is controlled discretely (e.g. Substance quantity is extremely small).
- Substance quantity is defined directly (e.g. Reactors determining the beginning of transcription, Reactors that divide all Substance quantity by two during cell division).
- Reactions calculated with algebraic equations (e.g. Reactors calculating Rapid Equilibrium Equations, Reactors calculating osmotic pressure).
While Normal Reactors use velocity calculations in determining Substance quantities, Postern Reactors manipulates all Substance quantities without considering a kinetic event. Thus Postern Reactors implement calculation results using the SetQuantity(Float q) method instead of the velocity (Float v) method.
Postern Reactors are generally named with a Class name, followed by " PReactor " (e.g. ABCPReactor).
Note that only one Substance can be referred to in each Postern Reactor. Substance quantities may not be controlled by multiple Postern Reactors. Accurate simulation cannot be guaranteed if more than one Postern Reactor is used to specify the reaction of one Substance. In cases where multiple reactions govern the quantity of a Substance, each of these reactions must be specified within one Postern Reactor (e.g. the GeneralRapidEquilibriumPReactor).
2 Reactor Description Files
A set of standard Reactors for commonly-used reactions are provided as a part of E-CELL2 (see Section 7 of this chapter). However, for more complicated or specialized modeling tasks, it may be necessary for the user to write RD files to create or customize Reactors.
2.1 Writing Reactor Description files
Each line of an RD file begins with a keyword, which is followed by an associated value (some keywords may require multiple values). There are several simple rules which apply to all RD files:
- The rest of a keyword comprises of capital letters and the " _ " character, and may not include any whitespace " " .
- All keywords begin with the " @ " or " % " character. Keywords beginning with " @ " require a single (scalar) value, whereas keywords beginning with " % " require a list of values delimited by commas.
- The rest of a keyword comprise of capital letters and the " _ " character, and may not include whitespace " " .
- A " # " character indicates that the rest of that line contains comments, and should be ignored by the converter. If a " # " character is to be placed at the beginning of a line, it must be preceded either by a backslash, " \ " , or whitespace. However, if " # " is not in the beginning of a line, it is outputted as it is.
- Lines that do not begin with a Keyword will be interpreted as a continuation of the previous line.(put " \\ " where you want to start a new line when converting into Spec Sheet.)
- Some keywords can be omitted, depending on the specific purpose of the RD file (see Table 1).
Keywords for General Information
@CLASSNAME:@BASECLASS:@AUTHOR:@EMAIL:@DATE:%VERSION:@BRIEF_DESCRIPTION:
The class name of the Reactor is specified in the
line beginning with the keyword, @CLASSNAME.
Note that the name of
the Reactor Description file must be CLASSNAME.rd.
The name of the base class that is inherited by @CLASSNAME is
specified in the line of the keyword, @BASECLASS. Most classes
inherit the base class, " FluxReactor " .
The name of the author of the
Reactor, their E-mail address, and the
date the Reactor was created are specified in the
lines of the keywords, @AUTHOR, @EMAIL and @DATE, respectively. There is no strict rule in the form of the date.
The version of E-CELL2 that is currently in use and
the version of this Reactor, separated by a comma,
are written in %VERSION line (e.g. ecs-v1.0).
The value for @BRIEF_DESCRIPTION
should be a one-line description of the Reactor.
These keywords will be read into the main E-CELL2 application.
Keywords for the Reactor Spec Sheet
@DESCRIPTION:@EQUATION:%SUBSTANCE:@NOTES:
The line @DESCRIPTION provides a detailed explanation of the
Reactor.
The line @EQUATION requires the equation of the Reactor
to be written in LaTeX displaymath mode. There are three ways of
writing in displaymath mode:
- $$ equation $$
- \begin{displaymath} equation \end{displaymath}
- \[ equation \]
Information regarding the Substances involved in the
reaction is specified in lines beginning with the keyword,
%SUBSTANCE. Four types of reaction-associated
Substances are pre-defined: Substrate, Product,
Catalyst, and Effector. It is also possible for users to create
their own customized Substance types.
The name of the
Substance, the maximum quantity, the minimum quantity,
and a short comment, all divided by a comma, is entered in the
%SUBSTANCE line. Numerical values are required in
these areas except when there is no limit to the number of
Substance types needed to perform the reaction,
in which case the maximum or minimum quantity value should
be set to " Inf " . Brief comments on the reactants can be written as
the last of the comma separated values for the %SUBSTANCE
line when a special Substance is defined.
These comments will appear only in the Spec Sheet.
General comments or warnings about the Reactor
can be written in the @NOTES line.
Keywords for the Reactor Source Code
%PARAMETER:%INCLUDE_FILE_H:@PRIVATE:@PROTECTED:@PUBLIC:%INCLUDE_FILE_C:@INITIALIZE_FUNC:@REACT_FUNC:@OPTION_C:
The %PARAMETER line defines the parameters, or arguments, that the
Reactor requires. In each %PARAMETER line, the name
of the parameter, the parameter type, the unit, and a description of
the parameter must be given as a comma-separated list. If the
parameter can only have an integer value, it is of type " Int " .
If the parameter has a non-integer value, it should be specified
as type, " Float " .
These type definitions are unique to E-CELL2 and should not be confused with the definitions of " int " and " float " used in C++ and other programming languages.
In E-CELL2, " Int " refers to a
32bit signed integer, while " Float " refers to a 64bit floating point number for the GUI mode, and an 80bit floating point number for the batch mode in a i386 compatible CPU.
The %INCLUDE_FILE_H line specifies other
Reactor Source Code files (.h) that are to be
included in the source code header file (.h).
File names listed here should not be contained in characters such
as <, >, or ".
Contents of the @PRIVATE,
@PROTECTED and @PUBLIC lines are
private, protected and public members respectively.
The %INCLUDE_FILE_C line specifies other
Reactor Source Code (.h) files that are to be
included in the source code .cpp file (.cpp).
File names listed here should not be contained in characters such
as <, >, or ".
If the base class FluxReactor is inherited, files
Reactor.h, Reactant.h, Stepper.h, and RootSystem.h has already
been included, in which case specification of other
Reactor Source Code files is unnecessary.
Note that Stepper.h, which existed in the E-CELL1 System, was abolished in the E-CELL2 System.
In cases where the FluxReactor
is not inherited, it is recommended to include the file
StandardHeaders.h. By doing so, the three header files will be
included in your Reactor automatically.
New methods and other macros are defined in the line, @OPTION_C.
Contents of @OPTION_C will be included in the beginning of its
corresponding .cpp file.
Initialization processes which are to be executed just once after Rule File is loaded and before
the simulation begins can be specified in C++ code in the @INITIALIZE_FUNC
line. Checking of parameter ranges and precomputation of constants
are typical initialization processes.
Processes to be executed for every simulation step should
be defined in the @REACT_FUNC line. For most
Reactors,
the two processes that need to be defined here are:
(1) computation of the reaction rate;
(2) increase/decrease of Substance quantities
according to the reaction rate in normal Reactors.
If the class inherits the base class FluxReactor, writing the code for (2) can be simplified by using the process() method. An argument of a process() method is the reaction velocity per second, and is a Float type argument.
Note that activity must be given as the activity value of 1 step, but this is recalculated and will be expressed as the activity value of 1 second in user interface such as the Reactor Window.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table1: Summary of Keywords (Applicable Files and its syntax)
|
(1)Included as comments
(2)Unnecessary for certain kind of Reactors. |
|
||||||||||||||||||||||||||||||||||||||||||||||
Table2: Summary of Methods
2.2 Examples of a Reactor Description File
The box below shows an example of a Reactor Description file (RD file) for a MichaelisUniUniReversibleReactor with the the following equation:
| v = | (KcFKp[S] - KcRKs[P])[E] | |
|
||
| Ks[P] + Kp[S] + KsKp |
@CLASSNAME:MichaelisUniUniReversibleReactor
@BASECLASS: FluxReactor
@AUTHOR: Kouichi Takahashi
@EMAIL: shafi@sfc.keio.ac.jp
@DATE: 1999 2/22
%VERSION: ecs-v08, 0.1
@BRIEF_DESCRIPTION:Simple Henri-Michaelis-Menten UniUni Reversible kinetics.
@DESCRIPTION:Simple Henri-Michaelis-Menten UniUni Reversible kinetics.
@EQUATION:$$v=\frac{(K_{cF} K_p [S]-K_{cR} K_s[P])[E]}{K_s[P]+K_p[S]+Ks Kp}$$
%SUBSTANCE:Substrate, 1, 1
%SUBSTANCE:Product, 1, 1
%SUBSTANCE:Catalyst, 1, 1
%SUBSTANCE:Effector, 0, 0
%PARAMETER: Ks, Float, mol/l, Michaelis Constant of Substrate
%PARAMETER: Kp, Float, mol/l,Michaelis Constant of Product
%PARAMETER: KcF, Float, mol/l, Catalytic Constant (Forward)
%PARAMETER: KcR, Float, mol/l,Catalytic Constant (Reverse)
@PRIVATE: Float Ksp;
@INITIALIZE_FUNC:
Float Ksp = Ks * Kp;
@REACT_FUNC:
Float S = substrate(0)->concentration();
Float P = product(0)->concentration();
Float E = catalyst(0)->quantity();
Float velocity = (KcF * Kp * S - KcR * Ks * P) * E / (Ks * P + Kp * S + Ksp);
process(velocity);
|
Values of Ks and Kp are constants, therefore, the product of Ks and Kp (Ks*Kp) will
also be only constant. This calculation need executed once at the
beginning of the simulation. This precomputation can be accomplished by
adding the following equation in the @INITIALIZE_FUNC line:
Ksp = Ks * Kp;
In the @REACT_FUNC line, a new variable, S, is defined to store the
concentration of the substrate. The concentration of the Substrate
is given by " substrate(0)->concentration() " , therefore:
Float S = Substrate(0)->concentration();
The value for P and E are also obtained by the same method.
3. Converting into Reactor Source Code (.cpp.h files)
The RD files must be converted into Reactor Source Code for the E-CELL2 System. This is accomplished by using the program,
" rd2ch.pl " in the directory " tools\reactor " . By standard installation, this directory is " C:\E-CELL2\tools\reactor " and " /cygdrive/c/E-CELL2/tools/reactor " on Cygwin.
By executing this program, a .cpp file representing
the source code, and a .h file, are created.
In the E-CELL2 System, a Reactor Source Code requires the " .cpp " suffix,
by default. It can be specified with the " -e " option.
Otherwise the user must change the suffix of an output to the correct one,
from the Window Explorer .
On Cygwin, use " cd " command to move to the directory where " rd2ch.pl " exists. The RD file is supposed to exist in this directory. To run the program, type:
% ./rd2ch.pl filename.rd
The output directory can be specified with the -d option,
% ./rd2ch.pl filename.rd -d output_directory
The directory where template file exists can be specified with the -t option. If this is not specified, the directory will be " template " , which exists under the directory where " rd2ch.pl " is.
% ./rd2ch.pl filename.rd -t template_directory
The suffix of a Reactor Source Code must be specified with the -e option. The source code file with " .cpp " is to be generated because this option is implicitly selected.
% ./rd2ch.pl filename.rd -e file_extension
Help is displayed with the -h option,
% ./rd2ch.pl -h
4. Converting into a Reactor Spec Sheet
The RD files can be converted into a Reactor Spec Sheet by using the program
" rd2tex " , which exists in the same directory as " rd2ch.pl " . Here in this manual, RD file is supposed to exist in the same directory as " rd2tex.pl " .
To run this program, type:
% ./rd2tex.pl filename.rd
As this program is executed, the number of keywords read by the system is displayed. Multiple input files can be specified at the command line, in which case a Spec Sheet which contains information on multiple Reactors is created in a single LaTeX file. Wild cards can also be used for specifying file names. For example:
% ./rd2tex.pl *.rd
The above example will create a list of all RD files in the current directory.
The default name for the output LaTeX file is
" ReactorDescription.tex " .
The -s option forces rd2tex to use
the stem of the input file name with the suffix " .tex "
(e.g. " filename.tex " for an input file named
" filename.rd " ). This can
be done when the conversion involves only one input file. When
multiple RD files are being converted into one Spec Sheet, the output
LaTeX file name can be specified with the -o option.
% ./rd2tex.pl filename1.rd filename2.rd.......-o output_filename.tex
Help is displayed with the -h option.
% ./rd2tex.pl -h
5. Creating Reactor Shared Objects and Spec Sheets using Makefiles
A C/C++ compiler will be needed to create Reactor. Refer to Chapter 1 for the detail description of installation.
Borland Borland C++ Compiler 5.5 gratis down load procedure
In the E-CELL2 System, Borland C++ Compiler 5.5.1 by Borland Software Corporation is used. The E-CELL2 system for standard Reactor model is installed in " C:\E-CELL2\standard " by default. This directory is recognized as " /cygdrive/c/E-CELL2/standard " on Cygwin. Note that RD file or source code of a Reactor for human erythrocyte is not included in this distribution.
There are 3 subdirectories, under " C:\E-CELL2\standard " , which are related to creating and using Reactor; " SRCR (hold source code of Reactor and header files) " , " DLLR (hold Reactor itself (.dll) for the GUI mode) " , " DLLRB (hold Reactor itself(.dll) for the batch mode) " .
To compile a Reactor, its Reactor Source Code and header file, generated according to the previous procedure, must be placed in the SRCR folder first. Then add the file name to create with " .dll " suffix to DLLFILESR part of Makefile, which is located in DLLR directory or DLLRB directory. By adding " \ " to the tail of the sentence, description of DLLFILESR over multiple lines can be made.
|
On Cygwin, move to the directory, for the E-CELL2 standard-reactor model, which is " C:\E-CELL2\standard " in standard installation, and " /cygdrive/c/E-CELL2/standard " on Cygwin. Here is a batch file " buildr.bat " . This batch file is used for creating DLL files and putting them into the appropriate directories. Before executing " bulidr.bat " , it is needed to open it with editor and substitute the underline part of " set PATH=c:\Borland\bcc55\bin;%PATH% " with absolute path of " bin " , which is located under the directory where Borland C++ Compler is installed.
After this operation is done, " buildr.bat " batch file should be executed on Cygwin. Type:
% ./buildr.bat
An object file(.dll), resulting from compilation, will be placed in the DLLR folder for GUI mode, and in DLLRB folder for batch mode.
To remove all the DLL files of Reactor, move to the DLLR directory on Cygwin, then type:
% make clean
Note that do not use GNU make.exe in Cygwin but Borland make.exe in Borland C++. These are not compatible. Be careful with PATH.
6. Running E-CELL2
It is necessary to keep in mind the following for normal E-CELL2 execution. A Reactor (.dll file) which is specified by RuleFile, must be in the Reactor directory (in standard configuration, DLLR and DLLRB are specified in the PATH of the starting-up batch file " ECELL2.BAT " and " ECELL2BB.BAT " ). A created Reactor dll file will usually be placed in these directories by " buildr.bat " . If E-CELL2 cannot find a specified Reactor or cannot find the PATH, an error message will be displayed.
7. Reactors provided by the E-CELL project
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table3: Standard Reactor
 is required to look at PDF.
is required to look at PDF.

|
E-CELL2 User's Manual Last Update $Date: 2002/07/16 16:45:42 $. |
Chapter 5 Creating User Defined Reactors [ Chapter 1 : Chapter 2 : Chapter 3 : Chapter 4 ][ Top ] |