- In early October, 2021, PDB files predicted by
AlphaFold Colabs
have no
header section,
making FirstGlance
unable to detect their provenance automatically. When a model has no header section
(and when no value in the
temperature column
of the PDB file exceeds 100),
FirstGlance now asks the user whether it is a prediction from AlphaFold.
In contrast, models downloaded from the AlphaFold Database have headers, and FirstGlance can identify these automatically. AlphaFold predictions processed by ConSurf are similarly handled.
Examples:- AlphaFold Colab prediction with no header section: Secreted in Xylem 1 of Fusarium oxysporum (Q709E2). You will be asked whether this is an AlphaFold prediction.
- AlphaFold Database prediction with header section: Iron phytosiderophore transporter of Zea Mays (Q9AY27). FirstGlance automatically determines that this is an AlphaFold prediction.
- AlphaFold Colab prediction processed with ConSurf: Secreted in Xylem 1 of Fusarium oxysporum (Q709E2). You will be asked whether this is an AlphaFold prediction. Includes putative ester crosslink (actually a clash).
- AlphaFold Database prediction processed with ConSurf: Iron phytosiderophore transporter of Zea Mays (Q9AY27). FirstGlance automatically determines that this is an AlphaFold prediction.
- Models predicted by
RoseTTAFold are now automatically recognized, and colored according to
RoseTTAFold's estimated error of position in Å. Examples:
- RoseTTAFold prediction for a Fusarium oxysporum protein of unknown function: RoseTTaFold prediction for UniProt W9JUF2.
- ConSurf analysis of the above RoseTTAFold prediction.
- The initial cartoon view of AlphaFold-predicted or RoseTTAFold-predicted structures is now colored by estimated reliability or error, respectively. A color key and explanation is displayed automatically, instead of the usual "Introduction" help. Try any example above (except ConSurf examples). For a ConSurf example, after moving to the FirstGlance control panel, go to the Views tab and click Error Estimates.
-
Instructions were provided
for submitting a sequence to an AlphaFold Colab to get a predicted structure.
These are linked at the
PDB code entry page, and in the Resources tab.
Improved:
- Electron density maps can now be displayed out to 15 Å (previously only to 3 or 5 Å).
- In the Crystal Contacts display (Tools tab), the contacting residues, initially
magenta, can now be displayed in white, or hidden.
Bugs fixed:
- A PDB file with (i) no header section and (ii) one or more protein crosslinks other than disulfide bonds, when (iii) loaded with FirstGlance in "more details" mode caused FirstGlance to remain permanently busy without ever populating the upper and lower left panels.
- For ConSurf results, the number of residues with incomplete sidechains was counted, but S- labels for missing sidechains were not applied when moving from the ConSurf control panel to the FirstGlance control panel. S- labels are still not applied in the ConSurf view until the user moves to the FirstGlance control panel.
- Links to a video introducing FirstGlance with its design goals were not working.
- When the model was uploaded (rather than specifying a PDB ID), saving an animation for PowerPoint failed.
- Using the Ligands+ or Water buttons made secondary structure beta strands lose their default thickness. (Technical: See hermite.txt.)
- Clicking on Find.. while viewing Crystal Contacts (Tools tab) hung FirstGlance in an infinite alert loop.
- When sidechains were hidden in the Vines/Sticks View, animating alternate locations failed to show sidechains. (Technical: sidechainsVisible; makeAltlocHelp().)
- Yellow halos disappeared when animating alternate locations. (Technical: see Jmol function colorByConfigs() in util.js.)
- When a homology model from Swiss-Model was uploaded, clicking Local Uncertainty left a BUSY indication in FirstGlance.
- After hiding hydrogen, using Disulfides/S/Se (Tools tab) showed a distracting and inappropriate alert.
New:
The only changes from version 3.51 are adaptations to handle structure prediction PDB files generated by AlphaFold, and to make users aware of AlphaFold and RoseTTAFold as sources of useful theoretical models. Thanks to Elsbeth Walker for bringing this need to my attention. Example: AlphaFold prediction for iron-phytosiderophore transporter yellow stripe 1.
- Chain Details and Sequences links are available in the Molecule Information Tab.
- In the Views tab, the "Local Uncertainty" link is replaced by a "Reliability Estimates" link, which, unlike the former, is available regardless of the fewer or more details mode.
- AlphaFold places reliability values in the temperature column, whereas empirical temperature values signify uncertainty. Thus, the colors are reversed for AlphaFold (blue for high reliability values, red for low reliability values). An appropriate color key is displayed.
- Touching an atom, when in "more details" mode, reports its estimated reliability.
- Clicking an atom, when in "more details" mode, reports its estimated reliability.
- AlphaFold is now recommended on the entry page and in the Resources tab.
- "Where do I find a PDB code?" on the
entry page
now includes instructions for AlphaFold and RoseTTAFold.
Bugs fixed:
- PDB files downloaded from the AlphaFold Database most regrettably failed to display in FirstGlance. (Although these PDB files contain only one model, it is identified unconventionally as "MODEL 0" rather than "MODEL 1". Technical: see load.txt.)
- When an mmCIF file was uploaded to FirstGlance, the model was displayed, but not all functions would work correctly. Uploaded files with filenames ending in .cif, or in mmCIF format (even if the filename ends incorrectly in .pdb) are now intercepted with appropriate error messages. (Technical: see isCIFFile and makeBadPDBFileHelp().)
New for AlphaFold Predictions:
The only change from version 3.5 is that the links to display electron density maps (or EM density maps) are visible in both simplified/"fewer details" mode and "more details" mode. The documentation below for version 3.5 has been updated to reflect this.
Thanks to Bob Hanson for implementing powerful, user-friendly density map commands in Jmol both for X-ray and EM maps, and thanks to Sameer Velankar for advice, and to PDBe at the European Bioinformatics Institute for providing a straightforward and efficient interface for fetching specified portions of X-ray and EM density maps. Thanks to Rachel Kramer Green for advice, and to RCSB.org for advice and for providing an ever-more powerful Advanced Search. Thanks to Ángel Herráez, Craig Martin, and Gabriel Pons for suggestions that improved this version, to Joel Sussman for advice and encouragement, and to Amr A. Alhossary for identifying some protein crosslink examples. Thanks to Jaime Prilusky's OCA Browser that makes it easy to find data entries with specified record types.
The previous FirstGlance remains available for comparisons: FirstGlance version 3.0.
- FirstGlance now starts in a simplified "fewer details" mode, showing less detail in the
Molecule Information Tab,
hiding some of molecule-specific information in the
Introduction,
and offering fewer options in the Views Tab and Tools Tab.
 Links
Show more details/views/tools
toggle visibility of more, becoming
Show fewer ....
There are three independent toggles, one for each tab. Details in the
Introduction toggle together with those in the
Molecule Information Tab.
Whichever modes you use last are automatically saved as preferences for your next session.
Links
Show more details/views/tools
toggle visibility of more, becoming
Show fewer ....
There are three independent toggles, one for each tab. Details in the
Introduction toggle together with those in the
Molecule Information Tab.
Whichever modes you use last are automatically saved as preferences for your next session.
Because FirstGlance in Jmol is the only macromolecular visualization program that indicates missing residues and incomplete sidechains in its initial view, and because awareness of both is important even for beginners (e.g. when charges are colored), these indications remain visible in the simplified/fewer details mode. Although the count of incomplete sidechains remains visible in the Introduction, it is hidden in the Molecule Information Tab.
FirstGlance may also be the only macromolecular visualization program that automatically detects unusual covalent protein crosslinks. The existence of any of the unusual crosslinks triggers an alert, even in "less details" mode, and even when "Alert for unusual components" is unchecked in the Preferences tab. Example: 2xi9.
HIDDEN in the simplified/fewer details mode:- Alerts for unusual components or features. (The previously existing preference for these alerts is turned off.) Example: 6cty. If you toggle from fewer to more details, these alerts will be shown.
- Molecular Views:
- Atoms with alternate locations. Simple example: 3hyd.
- Hide Dialog (in the Focus Box):
- Alternate locations checkbox.
- When hiding is inverted, the automatically-hidden alternate locations remain hidden.
- Molecule Information Tab:
- The R value. (Free R and its interpretation remain visible.)
- Count of non-glycines lacking beta carbons. Example: 1ijw.
- Count of incomplete sidechains. Example: 1ijw. (S- labels and count in Introduction remain visible.)
- Count of putative isopeptide bonds. Example: 3htl.
- Count of putative cysteine thioester protein crosslinks. Example: 2xi9.
- Count of putative cysteine thioether protein crosslinks. Example: 6e87.
- Count of putative threonine (or serine?) ester protein crosslinks. Example: 4mkm.
- Count of putative histidine-tyrosine protein crosslinks. Example: 1v54.
- Count of possible Lys-Cys NOS bonds. Example: 6zx4.
- Alternate locations. Simple example: 3hyd.
- Occupancies. Example: 1ijw.
- Link to view the PDB file.
- Deposition date.
- Related PDB entries. (New in this version.) Example: 1ijw.
- Introduction:
- Count of alternate locations.
- Views Tab:
- The Local Uncertainty color scheme.
- Tools Tab:
- Crystal contacts tool.
- Unit cell tool.
- Advanced/Technical.
- Alternate location, occupancy, and temperature (B factor) when you touch (hover over) or click an atom.
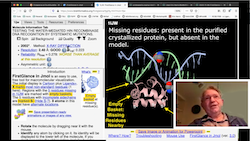
- A new video includes an introduction and the design goals for FirstGlance in Jmol. Links to the video are on the entry page, in the How & Where Index, and in the Introduction panel displayed with the initial view of a molecule.
New
|
Protein Crosslinks of several types (beyond disulfide bonds)
are now detected and reported.
An alert for
Unusual Components or Features
reports these regardless of preference settings.
Counts of each type present are linked in the
Molecule Information Tab,
subject to a preference setting.
A new tool "Protein Crosslinks" the Tools tab is always available.
Clicking one of those links/tools
lists the individual bonds of that type. Clicking on an individual bond finds it (yellow
halos), zooms in, and labels it and its neighbors. Also in the Lower Left Panel
are listed a range of examples, and the
methods used to detect the crosslinks.
The
methods are also listed in the Notes.
Only models with resolution of 4.0 Å or better are evaluated for protein crosslinks. When alternate locations are present, only conformation 1 is evaluated. When there are multiple models, only model 1 is evaluated. For most models, scanning for protein crosslinks adds no noticeable time to how long FirstGlance takes to stop being busy while initially characterizing the model. The median entry in the PDB has 3,600 non-hydrogen atoms. 90% of entries have < 13,000 non-hydrogen atoms. Comparing FirstGlance 3.0 vs. 3.5, ready time for 13,000 atoms (6RD5) is 9 sec for both. For 53,000 atoms (largest 1.5% of PDB, 6cri), ready times were 20 vs. 23 sec, respectively. For 99,600 atoms (largest 0.7% of PDB and maximum number of atoms compatible with the PDB format, 3J9Q), ready times were 34 and 37 sec, respectively. (PDB analysis: August, 2021. Timings: Chrome browser in Windows 10.) A single example is given for each type of crosslink below. More examples are listed under each type of crosslink in FirstGlance: click Protein Crosslinks in the Tools tab.
|
-
X-ray diffraction
electron density maps
and
Electron microscopy density maps
(example: 7dy0)
can now be visualized. They appear in a new browser tab. X-ray maps
are offered in both all-features maps (2Fo-Fc) and difference maps (Fo-Fc).
New in the Tools tab is Electron/EM Density Map
that explains:
Links for viewing maps are offered in three locations:

- In the Find.. dialog (Focus Box) after specified atoms have been identified with yellow halos.
- In the Ends.. dialog (Molecule Information tab/Tools tab) after one end of a chain has been displayed.
- For Protein Crosslinks (Tools tab), after one crosslink has been displayed.
Characteristics of the density maps viewer:- Extensive introductions and explanations are linked.
- FirstGlance determines whether the map is available before attempting to display it. Example: the map data for 1d66 were not deposited in the PDB. The PDB began requiring deposition of maps with models in 2008.
- The map isomesh can be shown within 3 or 5 Å of the target atoms.
- For X-ray models, the electron density map can be displayed for all features (2Fo-Fc), or for differences between the map and the atomic model (Fo-Fc).
- The target atoms are shown as sticks within the map. A checkbox displays them as balls and sticks.
- Controls are offered for slab thickness.
- The target atoms are colored by chemical element, with options to color them by absolute or relative temperature/B factor.
- Static images or presentation-ready animations (example at right) can be saved. See examples of static images for all-features and difference maps at Electron Density Maps in Proteopedia. More animations are in the slides that illustrate various animations saved from FirstGlance: tinyurl.com/movingmolecules.
- The authors' remarks
about sequences (REMARK 999) are now displayed at the top of the Lower Left Panel
when the Sequences link is clicked in the
Molecule Information Tab.
About 10% of the entries in the PDB contain REMARK 999. Examples:
3ZZZ,
5XAA.
- Sequence differences (SEQADV)
between those in the experimental molecule
and UniProt are now displayed at the top of the
Lower Left Panel when the Sequences link is clicked in the
Molecule Information Tab.
About two-thirds of the entries in the PDB contain sequence advisory (SEQADV) records.
Examples:
- 5XAA has N-terminal acetylation (1 line).
- 3ZZZ has an N-terminal expression tag (missing; 6 lines).
- 4a00 has engineered mutations (3 lines).
- 3odL has engineered modifications of a non-standard residue (10 lines).
- 4adp has expression tags at both ends (both ends missing), and one engineered mutation with different sequence numbering in the model vs. UniProt (10 lines).
- 1zeL has cloning artifacts and a conflict (8 lines).
- 2e00 has an N-terminal expression tag (with coordinates), one engineered mutation, and one deletion (6 lines).
- 1CGQ has insertions (3 lines).
- 110L has sequence conflicts (3 lines).
- 2f0w has 4 engineered mutations and one variant (5 lines).
- 2dxd referenced a non-existent REMARK 999, but when reported to the wwPDB it was remediated. Now its SEQADV lists initiating methionines and expression tags (all missing coordinates; 6 lines).
- 2ffi has modified residues (Met to MSE = selenomethionine), cloning artifacts, and a C-terminal expression tag (mostly missing; 30 lines).
- 6pv7 has linkers, conflicts, and C-terminal expression tags (missing coordinates; 45 lines).
- Related PDB files,
specified by the authors in REMARK 900,
are now listed near the bottom of the
Molecule Information Tab only in the more details mode.
About two-thirds of PDB files specify related entries.
Examples:
1o00 (10 described),
4j18 (described as open or closed),
4RPZ (11 not described),
1d0a (2 described + 5 not described),
4b2b (287 mostly described),
2fzv or
4mym (1 related, not PDB),
7nrx (1 EMDB described),
6myx (3 PDB, 2 EMDB).
- Caveats
provided by the authors are now displayed (in both fewer and more details modes)
above and near the
Focus Box in the
Molecule Information Tab. About 1.5% of entries in the PDB contain
CAVEAT records. Examples:
- 1v00: chirality errors even with resolution 1.7 Å and Rfree better than average.
- 1aaQ: atomic clashes.
- 5uu5: improper peptide linkages.
- 5a0a: "long bond".
- Obsolete entries now generate an alert, after which (unconditionally)
the superseding entry is displayed.
Approximately 2% of PDB codes designate obsolete entries. Examples:
1num,
4atc,
5tkx,
6r8d.
Improved - FirstGlance now defaults to high Quality rendering when the model has <= 33,000 atoms (~95% of entries in the PDB), unless a new preference is unchecked. The Quality button in the Button Box has final control. Rotation is choppier in high quality mode. Rotation of the Secondary Structure view is especially choppy, so its help panel now offers a checkbox to toggle off "solid" rendering of beta strands, and suggests turning off Quality. Examples: 7AHL has 23K atoms (17K non-H); 4zuL has 32.6K atoms (no H); 1f8s has 34K atoms (no H); 1g3i has 45K atoms (no H).
- When saving a static image, you now have the choice of low vs. high quality. (Until now, static images were saved in high quality unconditionally.)
- When saving a static image or an animation, new help is visible for centering or sliding the molecule before saving.
- FirstGlance now detects when the PDB identification code designates a molecule that is available in mmCIF format but not in PDB format. This occurs in ~1% of the entries in the Protein Data Bank, namely, when there are >99,999 atoms or >62 chains. FirstGlance cannot yet process mmCIF format, but it now suggests a workaround. Examples: 5LEG (pilus), 5NP6 (ribosome).
- Center atoms with halos is a new option in the Find.. dialog (Focus Box).
- Center Atom.. (Focus Box) has been renamed to simply Center.. and now offers to center atoms with halos as well as centering all visible atoms.
- The atom count, in the Molecule Information Tab, is now a link. When clicked, it shows the counts of non-hydrogen and hydrogen atoms in various categories.
- When uploading your own PDB file from the entry page, your uploaded file is now encrypted (via https).
- For macromolecular models with no Header (e.g. ATOM and HETATM records only), information in the Molecule Information Tab has been improved. As a test example, here is 3HTL with everything but the ATOM and HETATM lines deleted.
- The Find.. dialog (in the
Focus Box)
puts yellow halos around the found atoms. When finding alpha carbons (*.ca) or nucleic acid
backbone phosphorus atoms (*.p), on some backbones or smoothed backbone traces,
the halos were too small to see, especially at high zoom levels. These halos
have been enlarged, and now remain visible at all zoom levels.
(Technical: spacefill has been added to backbone, trace, non-sheet
cartoon, & ssbonds.)
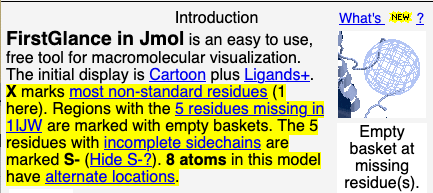
- In the Introduction, information specific to your molecule is now highlighted in yellow. Example shown at right: 1ijw.
- When non-glycine amino acids exist that are missing beta carbons, the count reported in the Molecule Information Tab (only in the more details mode) is now clickable, and will find those residues. Example: 1ijw.
- Text input form slots are now larger (with a larger font) in the Chrome browser, for example, the slots for entering the PDB code, and the Find slot.
- Examples were provided for nucleic acids with missing sidechains.
- In Firefox, initially-unchecked checkboxes are hard to see. They have been framed in gray
for the Vines/Sticks and Local Uncertainty views, help for the
Ligands and Water buttons in the Button Box, in the target selection dialog
for the Contacts tool.
Updated - Instructions for viewing all models in multiple model ensembles (example: 2bbn). The default viewer at RCSB.org changed, and does not offer a view of all models. By choosing the NGL viewer at RCSB, all models can be displayed.
- Web browsers compatible with FirstGlance in Jmol.
-
Mirror Sites for FirstGlance in Jmol.
Bugs Fixed - Under Sequences (Molecule Information Tab), the link to PDB-USA (RCSB) was broken. Thanks to Gabriel Pons for reporting this.
- Under Sequences (Molecule Information Tab), links to UniProt were missing for some protein chains. Example: 6byp. (FirstGlance was not obeying DBREF1, DBREF2 records for long UniProt accession codes.)
- The Secondary Structure view showed the backbone trace splitting as it entered, or coiled around, the rockets that represent helices.
- When there are D amino acids, the Introduction no longer says they are all marked with "X". Example: 6CNU.
- Depressing the Slab button failed to cancel ongoing centering, hiding, isolation, etc.
- Using the Zoom arrow buttons failed to cancel ongoing centering, hiding, isolation, etc.
- Toggling the Quality button uneccessarily canceled ongoing centering, hiding, isolation, etc.
*This version of JSmol incorrectly reports its release date as 2019.10.09.
-
New
- Salt bridges (Tools tab) can now be listed, spreadsheet ready. The listing includes every protein-protein pair of oppositely-charged nitrogen and oxygen atoms. It indicates which, if any, are backbone atoms at chain termini, and includes distances and various summary counts. An example of such a listing is 1hxw-salt-bridges.htm. FirstGlance does not list salt bridges involving nucleic acid or ligands+, but those can be visualized with Ends.. or Contacts & Non-Covalent Interactions.. (Tools tab).
- Contacts & Non-Covalent Interactions.. (Tools tab) has a new option to list counts of the contacting atoms in each category, as well as the counts of the contacted target atoms in each category. In addition, the listing includes terms that can be used with Find.. (Focus box) to apply halos to various subsets of contacting or contacted atoms. An example of such a listing is provided.
- The Molecular Mass of the model is now reported in the Molecule Information Tab on the same line as the atom count, just above the list of chemical elements. Clicking on MW Breakdown there reports molecular masses of components of the model.
- D-amino acids are now handled appropriately. There are >700 entries
in the PDB containing
one or more D-amino acids. Examples:
6cnu (small),
5e5t (larger),
2q33 (100% of non-Gly amino acids are D).

- D-amino acids are labeled "D", instead of the "X" used for other non-standard residues.
- The count of D-amino acids, and their percentage of non-Gly amino acids are reported in the Molecule Information Tab. The count of D-amino acids is also alerted at the outset as unusual components.
- Incomplete sidechains in D-amino acids are now marked S- and reported in the Molecule Information Tab. ("S-" supercedes "D"). Example: 5e5t.
- Charge (Views tab) now colors and counts D-amino acid charges correctly. Examples are given in the lower left (help) panel.
- Disulfides/S/Se (Tools tab) now handles Dcy-Dcy and Cys-Dcy disulfide bonds correctly. Examples are given in the lower left (help) panel.
- Salt Bridges/Cation Pi (Tools tab) now reports salt bridges involving charged sidechains of D-amino acids. Examples are given in the lower left (help) panel.
Handling of D-amino acids required many changes. Please let know if you run across something we missed. - A new Animation Kit has been provided for making presentation-ready animations of views in FirstGlance, in the event the built-in animation maker is unsatisfactory. See Animation Kit Instructions.
-
Instructions for aligning protein sequences, and displaying the results.
When you click Sequences in the
Molecule Information Tab, there is a link to these instructions
in the lower left (help) panel.
Improved - Salt bridges (Tools tab) at last include those involving charged N+ or COO- termini of protein chains. A link in the lower left help panel puts halos around all chain termini. An example of modest size that nevertheless includes several types of salt bridges including chain termini is 9ins. Further examples are provided in the lower left (help) panel when displaying salt bridges. Detection of salt bridges involving C-termini includes those in which the authors omitted one oxygen from the COO- terminus: see further information.
- Salt bridges (Tools tab) now displays counts of amino acids and atoms, and the percentage of amino acids involved in salt bridges (typically higher in thermophiles than in mesophiles).
- Cation-Pi Interactions (Tools tab) now include those involving charged NH3+ termini of protein chains. A link in the lower left help panel puts halos around all chain termini. Example: 6mt3 appears to have a cation-pi interaction between the Phe1 amino terminal N+ of peptide chain B and Trp167 in chain A. Note that also the opposite face of Trp167:A interacts with Arg170:A (deemed energetically significant by CaPTURE). Don't be confused by the interaction of the aromatic ring of Phe1:B with Arg62:A (also deemed energetically significant). Note that the CaPTURE server does not report the energetics of cation-pi interactions involving amino terminal cations. (An option to list cation-pi interactions was not provided because CaPTURE does this better.)
- Cation-Pi Interactions (Tools tab) now lists counts of cationic and aromatic amino acids engaged in putative cation-pi interactions. When cationic protein chain amino termini are involved, the count is given in magenta. Examples are 6mt3 and 7ahl. (The CaPTURE server does not report cation-pi interactions where the cation is an amino terminus.)
- The code that detects Cation-Pi Interactions (Tools tab) is now more accurate. It excludes some residue pairs, or unpaired residues, that were previously displayed but in fact fail strict criteria (Lys or Arg cation within 6.0 Å of three alternate atoms in an aromatic ring of Phe, Tyr, or Trp). However, the new code is slow, so it is used only when the model contains <12,000 non-hydrogen atoms (>90% of models in the PDB). For larger models, the old less accurate code is still used by default. In such cases, however, a link in the lower left help panel offers to re-detect cation-pi interactions with the slower, more accurate method. (More here, and for technical details, see catpi.txt.)
- Salt Bridges/Cation-Pi Interactions (Tools tab): The controls in the lower left help panel are now much faster (switching from salt bridges to cation-pi interactions, coloring, and whether or not to display thin backbones).
- In the table displayed by Ends
(Molecule Information Tab and Tools tab),
carboxy termini modeled with only one oxygen are now flagged ¶ and an explanation
is provided. Example:
1bl8.

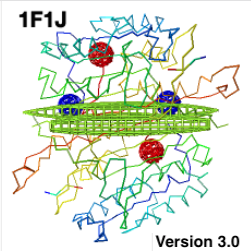
- Empty baskets representing gaps of missing residues within a chain are now
ellipsoids instead of spheres.
Missing residues at ends of chains continue to have spherical empty
baskets (diameter 6 Å).
The ellipsoids make it easier to tell which empty baskets represent within-chain
gaps. Also, when gaps are large, the former
huge spherical empty baskets obscured
the structure. An extreme example with 48 Å gaps is
1f1j, illustrated at right.

A case having 15 within-chain gaps with a wide range of gap distances is 6qj4. Here, the minimum within-chain gap distance between alpha carbons is 3.57 Å (Find: "362,366:c"). However, to ensure visibility, the minimum ellipsoid length is set to 7 Å. Thanks to Karsten Theis (Westfield State Univ., Massachusetts) for help with ellipsoids.
6oy5 has a DNA strand with ends missing, and a within-chain gap: Isolate DNA to see this clearly.
-
The dashed lines that indicate Distances/Angles (Tools tab) were becoming almost
invisible when the Quality button was depressed. Thanks to Bob Hanson for fixing
this issue in JSmol. (Technical: "quality" uses antialiasdisplay.)
- The
Molecule Information Tab
now says "No biological unit(s) are specified"
when there are none. It also says that multiple biological units are specified
when this occurs. The biological unit help now points out when "asymmetric unit" is not applicable
to the method. Test cases:
Asymmetric Biological Code Method Unit? Units* •1d66: X-ray Diffraction Yes 0 •1hho: X-ray Diffraction Yes 1 •1b6b: X-ray Diffraction Yes 2 (monomers) •5cev: X-ray Diffraction Yes 2 (hexamers, neither = asymmetric unit) •1qrd: X-ray Diffraction Yes 2 (software determined more likely than author determined) •5mny: Neutron Diffraction Yes 0 •2yz4: Neutron Diffraction Yes 1 •6pkj: Electron Crystallography Yes 0 •6oiz: Electron Crystallography Yes 1 •2bbn: Solution NMR No 0† •5v7z: Solid State NMR ? 0 •2c0x: Solid State NMR ? 1 •6ef8: Electron Microscopy No 0 •6nef: Electron Microscopy No 1 *Not counting the asymmetric unit, which is sometimes designated as a biological unit. † A search of the PDB finds no solution NMR entries with biological unit assembly operations (Sept. 14, 2019).
- Dipeptides
- Are now included in Ligands+ and in the initial view are rendered solid (spacefilling atoms). Previously, they were rendered as a tiny fragment of cartoon "coil", difficult to see, especially since they had the same chain ID and color as the chain to which the dipeptide is bound. In fact, the PDB considers dipeptides to be ligands: see policy and explanation which lists some examples.
- Also, now you will be alerted to the presence of dipeptides as unusual components.
- Also, amino acids listed under Ligands+ and Non-Standard Residues (Molecule Information Tab), when in dipepides, have a link to put halos on dipeptides.
-
Center Atom.. (in the
Focus Box) now offers two different ways to center all visible atoms.
In addition to the average position of the visible atoms
(previously the only option),
you can now center the boundbox
of the visible atoms. The help offers an example to illustrate the difference
(1zms). Thanks to
Ángel Herráez for help with this.
- A rough estimate is made of whether the biological unit may be too big for FirstGlance
(≳50,000 atoms). If so, the Biological Unit help recommends using the button
Get BACKBONE ONLY Biological Unit(s).
Example:
1sva (virus capsid).
(Technical: bioUnitTooBig.)
- Incomplete sidechains
(Molecule Information Tab)
is now clickable and puts
halos around the residues marked S-. This is particularly helpful in chains of D-amino acids.
- Checkboxes and radio buttons are larger. (Hours spent trying to give them more visible
borders ended without cross-browser success.)
- When a multiple-model PDB entry is loaded, the link View All Models in the
Molecule Information Tab
displays updated help, offering an external method to display all models.
Example:
NMR entry 2bbn.
- REMARK 400 is present in 4% of PDB entries. It contains additional information about
the material studied. It is now displayed underneath the per-chain information in the
Chain Details help. Its fairly amazing what you can find in there. Examples:
1vif,
5u66,
5cxt,
3L8L,
3Lqr,
3m9s,
209d,
2iwe.
Thanks to
OCA's Record Name search capability
for making it easy to find entries with REMARK 400.
- The Chain Ends (Termini) dialog is now accessible from the
Molecule Information Tab next to chain details and sequences.
- For cryo-EM, Local uncertainty (Views tab) now documents that temperature/B factor values are usually less meaningful than they are for X-ray crystallography.
- Additional examples have been provided in the lower left (help) panel for Secondary Structure (Views tab), as well as for Disulfides/S/Se and Salt Bridges/Cation-Pi (Tools tab).
- How do I ...? Where can I find ...? The Index to FirstGlance in Jmol has been expanded with dozens of new entries taken from Notes for FirstGlance in Jmol and All About FirstGlance in Jmol. Also, the latter two documents were updated.
-
Design & History of FirstGlance in Jmol was updated and greatly expanded.
Bugs fixed - The Charge.. view was failing to color C-terminal oxygens red on monomeric amino acid ligands (example 4ffL), or dipeptide ligands (examples 2cyh, 1dpp). To isolate these unusual ligands, Find "protein and hetero" and then Isolate atoms with halos. Also, these ligand C-termini were not being reported and counted in the Charge help panel. Since, by PDB policy, monomeric amino acids and dipeptides are considered to be ligands, and are given the same chain ID as the protein chain to which they are bound, their ends are not listed in Ends (Tools tab). In contrast, tripeptides (or longer peptides) are given distinct chain identifiers, and are listed in Ends (example: 4q1L). (The amino termini on mono- and dipeptide ligands were correctly colored blue before this bug fix. This bug did not affect polypeptides of length 3 and longer.)
- Standard amino acids in dipeptide ligands were being marked "X". (Technical: "and not amino" in selectNonStandardResiduesSpt.)
- For the Charge.. view, the counts of visible charges reported in the help panel (lower left) reported an incorrect number of visible charges on incomplete sidechains when some incomplete sidechains were hidden. The invisible ones were not being deducted from the counts.
- The ends of Dinucleotides were not being listed when requested with Ends. Examples: 3rtj (RNA); 6mdx (DNA). Dinucleotides are handled differently than dipeptides, the latter being deemed ligands by the WWPDB. See WWPDB policy.
- Cation-Pi Interactions (Tools tab) occasionally displayed an aromatic ring without an interacting cation partner, or vice versa. Example 1: Tyr6 was displayed without a partner in 4enl. Example 2: Arg141 in chains A and C were displayed without a partner in 2hhd. After a bug was fixed, neither of these is displayed. CaPTURE deems the interaction between Arg141:A and Trp37:D in 2hhd to be energetically significant, but the distance between those residues exceeds the 6.0 Å limit used by FirstGlance. CaPTURE deems Tyr6 in 4enl not to be involved in an energetically significant cation-pi interaction.
- After coloring by Local Uncertainty (Views tab), unhiding things (Hide.. in the Focus Box) failed to re-display them.
- Unhiding moieties hidden in the Disulfides/S/Se view (Tools tab) failed.
- After showing Contacts & Non-Covalent Interactions to atoms with halos, then using Find.. to put halos on something else, returning to Contacts Shown incorrectly reported the current atoms with halos as the target for the previously initiated Contacts analysis.
- A fragment of uridine-5'-monophosphate in 4dw5 was invisible in the initial view and failed to be displayed by the Ligands+ button. The fix should ensure that all amino acid and nucleotide monomers are handled properly,
- Structures determined by electron microscopy were labeled "asymmetric unit" in the Molecule Information Tab.
- Crystal Contacts and Unit Cell (Tools tab) were handled incorrectly for structures determined by electron microscopy.
- Crystal Contacts and Unit Cell (Tools tab) left JSmol blank while the computations were being completed.
- D-amino acids were not handled correctly. See item #4 above.
- Thin backbone traces were still almost invisible, when the Quality button was depressed, for Disulfides/S/Se.. and Salt Bridges/Cation-Pi.. (Tools tab).
- "BUSY" disappeared well before completion of the Salt Bridges/Cation-Pi view (Tools tab), and before coloring by Local Uncertainty appeared, and before the Disulfides/S/Se.. view (Tools tab) appeared.
- "BUSY" was not shown during the first request for Charges or Chain Ends or Alternate Locations, while the necessary information was being gathered. Also it was not shown while secondary structure was calculated by JSmol.
- While alternate locations are animating, the following operations failed: Center Atom, Reset, Zoom (from arrow buttons), Crystal Contacts, Unit Cell. Everything else seemed to work. Let know if you run across something we missed.
- When visualizing ConSurf results, the link to help about "insufficient data" was broken.
- After applying yellow halos with Find.., "Re-center all visible atoms" left halos on the wrong atoms.
- With biological units (assemblies) constructed with the MakeMultimer server, clicking on an atom reported multiple elements, some incorrect.
- Salt Bridges/Cation-Pi.. (Tools tab) failed to raise the Ligands+ button.
- The temperature reported after clicking on an atom sometimes had many digits after the decimal point. There should be only one.
- Displaying 2mcg as thin backbones, putting halos on PCA with Find, and isolating atoms with halos caused everything to disappear. (Technical: see "zoomto 1.0 {not ~hidden_jdef}".)
- 1w0e hung FirstGlance with a permanent busy condition. This was because Leu 261, which precedes a gap of missing residues, has only one atom, the main chain nitrogen. All other atoms were missing, and FirstGlance was looking for its alpha carbon upon which to place an empty basket. Thanks to Samantha Ziemniak (Manchester University, North Manchester, Indiana, USA) for reporting this bug.
- 6rt4 hung FirstGlance with a permanent busy condition when Ends were requested.
This was because chain C, a trinucleotide, has only one complete residue. The second is
missing all but phosphate, and the third is completely missing. The ends of chain C are now
reported as "not found", and chain C is included in "Ligands+". (Technical: see getEndsGIs().)
Java No More - FirstGlance version 2.74 was the last version that worked with the Jmol Java applet. Due to an unintentional change that has not been identified despite considerable effort, versions 2.8 and later do not work with the Java applet. This is not much of a problem since very few web browsers still support Java. The Preferences tab, and Notes have been updated to reflect this change. Let know if you run across something we missed. (The standalone Jmol Java application continues to work just fine.)
-
The new default rendering is Shiny, especially noticeable for solid views. You can go back to the old less shiny and slightly brighter rendering in the Preferences Tab. This information is announced the first time you click the Quality button.

New default "Shiny"
rendering.Older rendering,
now a preference option. - The number of locations/atom are now counted for each atom with an alternate location code. These counts are summarized in the lower left (help) panel when you click on alternate locations (in the Introduction help or in the Molecule Information Tab). The most common case is 2 locations/atom, each with 50% occupancy (e.g. 2uus), However, there are cases with 3 to 5 locations/atom (e.g. 1alx), and it is suprising how often atoms with alternate location codes have only 1 location (e.g. 2gn2). Thanks to Jaime Prilusky, we know that 42% of entries in the PDB have alternate location codes.
- Hide can now hide atoms with alternate locations, leaving only the
first conformation visible. For example, load
3hyd and click on alternate locations
in the Introduction (initial lower left panel) or in the Molecule Information Tab.
There, click on Show All As Sticks. Next, click on 2 to put halos around Glu3.
You will see that it has 2 conformations. Now go to Hide (link in the box at the bottom of
each of the 1st 3 tabs), and check "Alternate Locations". One conformation of Glu3
disappears.
- The first two alternate conformations can be animated. For example, load 2agt and click on alternate locations in the Introduction. Follow the "Suggestion", 3 steps at the top.
- The range of occupancy values is now reported under the atom count in the Molecule Information Tab. Separate ranges are given for atoms with vs. without alternate locations.
- A new occupancy report (click on Occupancy in the Molecule Information Tab: the report appears in the lower left "help" panel) lists all occupancy values occurring in the model. You can select atom locations with a given occupancy value by clicking on the value. From this report, you can generate a list of all atoms with partial occupancies.
- Ends (in the Tools tab) has a new option to Find all visible chain ends.
- There is a new preference checkbox for those few who would like to be alerted
when partial occupancies occur without alternate locations and a few other unusual situations.
Its default is unchecked.
Improved - Thin Backbones (Views tab) are no longer nearly invisible when the Quality button is depressed -- provided the zoom level is high enough.
- When you touch an atom, its hover report now always includes the atom's temperature. If the model has alternate locations or occupancies < 100%, those are also included. Ditto for the report below the molecule when you click on an atom.
- When a group/residue contains atoms with alternate locations, Hide and Isolate now act on only the clicked-upon alternate location, provided that you opt to hide only the clicked-upon group. If you opt to hide all such groups, all alternate locations will be hidden. For example, 1izh has alternate locations for the ligand. In fact, the ligand is modeled in two completely different but overlapping orientations.
- The cluster of buttons in the Views and Tools tabs has been labeled Button Box. This makes it easy to refer to in the "How & Where" Index.
- The cluster of controls that includes Hide, Isolate, Find, etc. in the Molecule Information, Views and Tools tabs has been labeled Focus Box. This makes it easy to refer to in the "How & Where" Index.
- An example has been provided that has multiple models and also atoms with alternate
locations. See item #22 under
Unusual components or features. Thanks to Jaime Prilusky and
OCA for making it possible to find such examples.
Bugs Fixed - Unchecking the Labels: Show checkbox failed to hide "X" labels when no sidechains are incomplete. Examples: 1bkx, 1k28.
- Hiding hydrogens was not remembered between the Hide.. dialog and the Vines/Sticks View.
New
(JSmol was not upgraded to 14.29.42_2019.05.01 because it does not work in Java mode.)
Version 2.8 is much faster at getting large models ready for interactive exploration.
It also has significant improvements and bug fixes.
It also has significant improvements and bug fixes.
-
Transfer of large
atomic coordinate files
(PDB files) through the Internet is very fast.
This is best seen with files too large for
the PDB format
(>99,999 atoms). Less than 1% of entries in the
Protein Data Bank
are too large for the PDB format. These are available only in the mmCIF format, supported by
Jmol/JSmol but not yet by FirstGlance.
- JSmol: 3j9q.pdb (99.6K atoms, 1.4 MDa) loads and displays in 7 sec. (load=3j9q.pdb)
- Jmol (Java): 5iv5.cif (550K atoms, 6.5 MDa) takes <5 seconds to come through the Internet, and displays in 9 sec. (load=5iv5.cif)
- JSmol: 5iv5.cif takes 70 sec to display.
The following results are for the Jmol Java application or JSmol alone (not FirstGlance):
After JSmol has loaded and processed the model data, FirstGlance takes additional time to gather data and analyze the model, before it displays the model ready for interactive exploration. As new capabilities have been added to FirstGlance in recent years, with larger models, the time between entering a PDB code and displaying the model ready for you to begin interactive exploration (the "ready time") kept increasing. This was because the data for all possible FirstGlance views and tools were being gathered before the model was first displayed, and because of a bug in code looking for cyclic nucleotides. The ready time became unacceptably long for models with >~33K atoms (MW >500,000), namely the largest 2% of entries in the PDB. To reduce the ready time, FirstGlance version 2.8 is ready before some data are gathered. Those data are gathered only if and when the relevant views are requested. Here are ready times for models of various sizes:
|
FirstGlance Version |
1d66 3K atoms3 28 kDa Smallest 26%7 |
7ahl
23K atoms4 233 kDa Largest 6%7 |
6cri
53K atoms3 886 kDa Largest 1%7 |
3j36 60K atoms3, 5 1.2 MDa Largest 1%7 |
3j9q
99.6K atoms3, 6 1.4 MDa Largest 0.7%7 |
| 2.8 |
0:04 (0:34)
0:03 (0:30) 0:03 (0:32) 0:08 (0:25) 0:17 (0:35) |
0:10
0:06 0:08 0:27 0:33 |
0:20
0:15 0:19 |
0:20
0:11 0:18 1:16 1:36 |
0:33
0:17 0:27 |
| 2.74 |
0:04
0:03 0:04 |
0:15
0:09 0:17 |
0:54
1:00 2:05 |
3:35
>7:00 >11:00 |
3:30
n.d. n.d. |
|
1. Min:sec between entering the PDB code and the final disappearance of "BUSY" after the tabs at
the top left had appeared.
Times measured after JSmol & FirstGlance were already in the browser cache
(1d66 had previously been displayed in the same browser session).
2. Edge and Internet Explorer operate JSmol unacceptably slowly. See
browsers.
Italics: First load (times measured after the browser cache had been cleared). Clocked on a mid-2014 MacBook Pro (Mojave, 2.2 GHz i7, 16 GB 1600 MHz DDR3). Internet Explorer and Edge clocked in a Windows 10 virtual machine (VMWare, same hardware). 3. There are no hydrogen atoms in this model. 4. Atom count includes hydrogens. Non-hydrogen atom count for 7ahl=17K. 5. Mostly RNA: 36K atoms RNA, 24K atoms protein. 6. 99,999 atoms are the maximum possible in the PDB format. FirstGlance does not yet handle mmCIF.
Greater than 99% of entries in the PDB are available in the PDB format.
7. Percentages of entries in the PDB determined January 23, 2019 by MW search,
when there were a total of 148K entries in the PDB.
|
|||||
- A new tool for javascript developers, Report Elapsed Times, is available at the
model specification page
by clicking Advanced Options. This was used to find the slowest parts of the code so
they could be improved or postponed.
Updated - Recommendations for web browsers that work optimally with FirstGlance have been updated. The changes in this version of FirstGlance enable more browsers to work well with it.
- The help for Disulfides/S/Se (Tools tab) has been updated: during remediation
of the Protein Data Bank, chemical identifiers CSE and CSW were updated to SEC and OCS
respectively.
Improved - In order to decrease the "ready-time" (see above) for larger models,
the following operations are no longer
performed before FirstGlance is ready for interactive exploration. They are postponed until the
dependent views or tools are requested, causing at that time a small delay for larger models.
- Secondary structure determination by JSmol. You can view the author-specified secondary structure immediately, but if you ask to view JSmol's objective secondary structure, there will be a small delay while the calculation is performed.
- Characterization of chain termini, needed for the Ends View. Locating the ends of chains and determining whether the terminal charges are missing is not trivial.
- Preparation for viewing charges. This includes characterization of chain termini and whether the charged terminal residues are missing or the terminal charges are blocked.
- When more than 100 sidechains are incomplete (missing sidechain atoms), the default label "S-" is no longer applied because of the clutter it causes. An alert explains the situation and options. Examples:
- The methods for determining chain termini ("ends") have been copied from a sourceforge email post into a local document.
- Ends (Tools tab) has a new example. 1HVZ is a cyclic 18-amino acid. Both ends are blocked by cyclization.
- Toggling spin, background color, quality, and slab are now faster for large molecules.
- When there are multiple models, clicking View all models (in the Molecule Information Tab) now provides a link to view all models at RCSB, since FirstGlance using JSmol loads only the first model. As before, another solution is to use Java with FirstGlance (See the Preferences Tab).
- When the query entered in the Find.. dialog is longer than the input box, the input box changes to a multiple line textarea with a submit button.
- When water is displayed with the Water Button after an Isolate operation has been performed, the report now gives the counts of waters hidden and visible.
- In the help for the Water Button, when you check Smaller Water, hydrogens are now shown as sticks. Formerly, they were balls the same size as the smaller water oxygen balls.
- "BUSY" is displayed more consistently when FirstGlance is working on the requested view.
- The help for Thin Backbone (Views tab) now tells you how to display a thicker backbone.
- There is now a thin gray border around the cluster of controls that is
at the bottom of every tab (except the last tab, Preferences).
This border is to emphasize that
those clusters are identical in the first 4 tabs.
Bugs Fixed - Detection of cyclic nucleotides had been coded inefficiently and was taking far longer than necessary, especially for RNA. Thanks to Angel Herráez for suggesting how to make the JSmol code much more efficient.
- The Charge.. view was failing to color C-terminal oxygens red when the C-terminal COO- was missing one oxygen (namely, missing the oxygen designated OXT in the PDB data format). Examples: 1bl8, 5zgg. 5yxb is an example having a C-terminal Lysine (chain B) with a missing sidechain; thus, the Charge.. view of the C-terminal residue of chain B is light blue with one red oxygen. (Most PDB entries have two oxgens on the C-terminus [.O and .OXT]. These were correctly colored red before this bug fix.)
- The report of Find was failing to count nucleic pentose C5' atoms when they were found in nucleotides that are not part of a chain. Examples: A351 in 1bkx; cyclic AMP "ACK1382" in 2ype. (Technical: see "define ~nucleic" vs ~polynucleic in scripts.js.)
- The report of Find was failing to count ligand+ atoms when they were found in amino acids or nucleotides that are not part of a chain. Example: Gly308 in 4cpa. (Technical: see foundLigandPlusCount in scripts.js.)
- When a query that cannot be interpreted (example: >1) was entered in the Find dialog, a permanent BUSY and other problems ensued. (Technical: fixed with clearFindForm() in consurf/msgcall.js)
- Rapid repeated clicking of the zoom arrows caused JSmol to freeze (no rotation).
- The Solid view was hiding hydrogen.
- Center All Visible Atoms (under Center Atom) was erasing empty baskets.
New
- The performances of popular web browsers with FirstGlance and JSmol, and their support for Java, have been updated. Firefox remains the best browser to use with FirstGlance for both Windows and Macs, primarily because it completes pre-processing javascript (before displaying the molecule) much faster than Chrome. The recommendations and warnings given by FirstGlance have been correspondingly updated.
- The list of
mirror servers has been updated.
Improved - The form slot and buttons for uploading a PDB file or entering the URL of a PDB file
are now larger and more readable
here.
Bug Fixed - The mechanism to Upload your own PDB file was failing intermittantly. Huge thanks to Jaime Prilusky for implementing a solution.
Updated
- The Copyright, Licenses, Acknowledgement page was broken.
- At the page where the molecule is specified, when using the option enter a molecule's URL, the ready-made example was not working.
Bugs Fixed
Now Secure (HTTPS): Since its inception in 2006, FirstGlance was using the old, insecure HTTP protocol. Peter C. Morris (Heriot-Watt University, Edinburgh UK) kindly reported a problem, which resolved to the fact that his University network required the newer, secure HTTPS protocol. More and more, institutions will be requiring HTTPS. Thanks also to Ingrid Barbosa-Farias who recommended the change to HTTPS. Thanks to Jeff Bizzaro and the team at bioinformatics.org, HTTPS was already supported by the server. FirstGlance has now been configured to use HTTPS regardless of whether the link/URL specifies the old HTTP protocol, and regardless of whether you start at index.htm or fg.htm. The PDB file is also now retrieved from the Protein Data Bank via HTTPS.
- When viewing a ConSurf result, clicking Go to the FirstGlance Control Panel removed the ConSurf coloring and changed spacefill to cartoon. The view should have remained unchanged.
- In Salt Bridges/Cation-Pi (Tools tab), when ConSurf Colors was checked, inapplicable radio buttons for other color schemes were visible.
Bugs Fixed:
-
Animations for presentations
are now generated externally (on a server).
Making a Powerpoint-ready animation now takes typically only 1-3 minutes, much faster than before,
and works in
all popular web browsers in Windows (all recent versions) or OS X.
Examples of animated slides:
tinyurl.com/movingmolecules.
Thanks to Jaime Prilusky, Weizmann Institute of Science,
for a great deal of help with this, and Angel Herráez, University of Alcalá, Spain,
for some of the code. Thanks to Jaime Prilusky and Joel Sussman, Weizmann Institute
of Science, for allowing FirstGlance to use the server of Proteopeda.Org for generation
of Powerpoint-ready animations.
- New: Ends (Chain Termini) in the Tools tab. End residues are listed,
along with whether the actual chain termini are missing.
Blocking and charges of protein ends are indicated.
Clicking on a terminal residue in the table zooms in
and shows it in atomic detail.
- New: Over 15 unusual components or features will now be announced in an alert
at startup.
Examples and methods for Find
are provided for each. All unusual components or features
can be haloed by clicking on the
 that appears below the molecule when unusual components or features are present.
This can be turned off with a new preference checkbox.
that appears below the molecule when unusual components or features are present.
This can be turned off with a new preference checkbox.
-
Java
is no longer encouraged as a checkbox option when specifying a PDB code, or in the preferences
of FirstGlance in Jmol. Chrome, Firefox, Edge, and Opera no longer support Java. Oracle
plans to stop providing Java plugins for web browsers. However, when using a
browser that supports Java, you can still invoke the Jmol_S signed Java applet.
After you have displayed a molecule in FirstGlance (using JSmol),
type &java onto the end of the FirstGlance URL in the browser address line, and then press
Enter.
Improved: - At the initial page, a help link has been added: "Where do I find a PDB code?".
- Secondary Structure (in the View tab):
- Now reports 310 and pi helices as well as alpha helices.
- The default display now uses secondary structure calculated by JSmol. Alternatively, radio buttons offer to show the authors' designations.
- When portions of the protein are hidden, percentages of each secondary structure element are now reported for only the visible protein. (Previously, the percentages were always for the entire model.)
- Charge (in the View tab) now reports whether the amino and carboxy ends of protein chains are charged, missing, or blocked. This includes end-charges for mono- and dipeptides, as well as longer polypeptides.
- When depressing the Ligands+ button, the help now states when no Ligands+ atoms are present in the model, or all are hidden.
- In Contacts & Non-Covalent Interactions (Tools tab), a new checkbox has been added to hide "empty baskets".
- Plans for teaching, learning, assessment have been linked into the Resources tab. These include syllabi for undergraduates and post-graduates, along with challenge questions for assessment reports.
- For PDB files lacking a header, the total number of atoms is now given, and chemical elements present are now listed, with a link to show the counts of each. See examples under Bugs Fixed #27.
- "Show H/non-H atom counts" was previously available only at the bottom of the hydrogen help (displayed by clicking on the hydrogen atom report in the Molecule Information Tab). It is now also available at the bottom of the help that shows counts of each chemical element (displayed by clicking on the count of different elements in the Molecule Information Tab). In addition, the display of that list, now titled "H/non-H Atom Counts by Composition", is now unconditional.
- When a PDB code fails to be retrieved from RCSB, the help now lists the possibility that the model is too large for the PDB format. Example: 5leg.
- The N->C Rainbow view of nucleic acids showed a backbone that did not extend beyond the phosphate of the 3' residue. This has been changed to a Jmol "trace" that extends beyond the phosphate of the 3' terminal residue to its 3' OH. (Both methods already extended to the 5' OH.)
- PDB-REDO has been added to the Resources tab.
- In the Gallery, the biological unit 1k28_mm1.pdb.gz has been updated for conformity with current PDB format requirements.
- When the model contains only alpha carbon atoms for protein, and/or only backbone phosphorus
atoms for nucleic acid, the user is
alerted unconditionally. Such models (rare) are still handeled poorly.
Bugs fixed: - The Charge view was reporting protein C-terminal charge only when both oxygens were present on the terminal carboxyl ("OXT" in PDB terminology). It turns out that quite often only one oxygen is present on the terminal carboxyl. Provided the C-terminus is not missing or blocked, its charge is now tallied regardless of the number of oxygens in the model. See also Charge under "Improved" above.
- The Charge view was not reporting N-terminal charge. This was a longstanding bug that was, as expected, complicated to fix due to the difficulty of identifying chain termini in a general way. See also Charge under "Improved" above.
- When no protein was visible, the Charge view was reporting zero charges, instead of stating that this view applies to protein only.
- The composition of a chain (Protein, DNA, or RNA) was occasionally being reported incorrectly under Chain Details (a link in the Molecule Information Tab). For example, the protein chains in 1flo were labeled DNA, and the RNA chains in 4wkr were labeled protein.
- Single nucleotides, not part of a chain of standard nucleotides, are supposed to be spacefilled in the initial view, as part of Ligands+. This failed for AMP in 5gz5, for cyclic-AMP (CMP) in 5kjx, and for cyclic-GMP (PCG) in 5c8w.
- After entering Isolate, and then clicking the button Cancel Isolate, clicking a chain hid the clicked chain instead of reporting the identity of the clicked atom.
- After entering Isolate and clicking one of the checkboxes (protein, DNA, RNA, carbohydrate), then clicking on a chain hid the clicked chain instead of reporting the identity of the clicked atom.
- After hiding a chain, residue/group, or atom, when next attempting to hide a range, after clicking the starting residue of the range, the radio button failed to stay on "Range" for the second click that would specify the end of the range.
- The Java applet (Jmol_S) was failing to load PDB files, showing a red message "script ERROR: unrecognized file format ...". This was due to the PDB changing pdb.org to go to wwpdb.org instead of rcsb.org. This fix went live in version 2.51 at bioinformatics.org on July 25, 2017.
- PDB files lacking headers were failing to load. Examples:
- Caffeine.
- Links using FirstGlance at Metal-Ligand Polyhedra (in Proteopedia).
- The count of hydrogen atoms was being reported as "undefined" when the PDB file lacked a header. See examples in the previous item.
- When hydrogen atoms are present, clicking on the hydrogen atoms report
in the Molecule Information Tab displays additional information about
hydrogen. An option at the bottom is Show H/non-H atom counts. This option is also
present at the bottom of the help panel listing counts of each chemical element,
obtained by clicking on elements in the Molecule Information Tab.
- When the PDB file lacked a header, this option was reporting "undefined" for water and non-water solvent.
- The count given for "non-H for ligand" included hydrogen.
- The report for clicking on an atom in amino acid UNK (unknown) was garbled. This was a bug in Jmol, kindly fixed by Bob Hanson. Example: 4o46.
Major:
-
Improved:
- When saving animations while using Java, sometimes the saving fails with "ERROR: capture canceled". A way to prevent this has been documented, and help now appears automatically when this condition arises. See Troubleshooting.
- When saving animations while using Java, sometimes the saved GIF file is empty (zero bytes). Suggestions for avoiding this have been improved.
- When using JSmol, PDB files are fetched from the Protein Data Bank (rcsb.org)
via a small program, jsmol.php. This program has now been implemented on the
bioinformatics.org server. Previously FirstGlance was using jsmol.php at stolaf.edu.
The change means that a rare out-of-service occurrence at stolaf.edu will not
prevent FirstGlance from working.
Bugs fixed: - Item #22 under Bugs Fixed for version 2.5 was not fixed correctly. Also, the bond not shown in the example given there was between the main chain N of Thr198 and the carboxy carbon of Asn197, not the bond with NAG697.
Version 2.5 enables saving images and presentation-ready animations.
-
New:
- Static images and animations can now be saved directly from FirstGlance in Jmol. Animations can be 360 degree rotations, or rocking, and will display as animations in presentations (e.g. Powerpoint, free Google Slides or Libre Office) or in web pages.
- Examples of animations saved from FirstGlance are provided:
- A summary has been provided of browsers compatible with saving images and animations from FirstGlance.
- FirstGlance can now be used to save a presentation-ready animation of any
molecular scene in Proteopedia.
Instructions are provided.
Proteopedia's easy-to-use Scene Authoring Tools (see the
Video Guide)
can be used to customize rendering and coloring, and the customized scene
can then be saved as a presentation-ready animation.
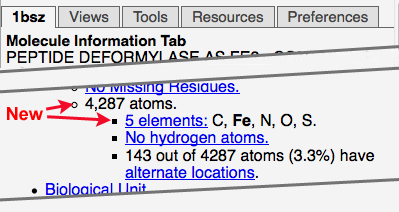
- The total number of atoms in the model is now reported in the Molecule Information Tab.
- The chemical elements in the model are now reported in Molecule Information Tab, with
a link that shows the counts of each element, and enables finding an element in the model.
- When the model includes selenium (Se), you are referred to Disulfides/S/Se in the Tools tab.
- When the model includes selenomethionine (MSE), a link is provided to an explanation. Example: 4hik.
Updated: - Documentation about using images or animations from FirstGlance in presentations has been updated.
- The introductory overview What Is FirstGlance? has been updated.
- The performance of JSmol in different browsers has changed, and the results of browser re-testing are summarized.
- The Opera browser no longer supports Java, so FirstGlance now blocks attempts to use Java in Opera.
- Installing and enabling Java has been updated.
- Enolase, an enzyme in glycolysis, colored by evolutionary conservation,
has been added to the
Gallery under ConSurf Examples.
Improved: - The help for Asymmetric Unit/Biological Unit now tells you if you are looking at the asymmetric unit. Thanks to Stacy Tchouangouem for this suggestion.
- In Contacts & Non-Covalent Interactions (Tools tab), in the help panel (lower left), the element color key has been moved up to a more visible position.
- Instructions for testing old versions of FirstGlance are now included
at the top of this document.
Bugs fixed: - Center Atom, when using Java, froze Jmol_S starting in FirstGlance 2.4. Thanks to Bob Hanson for help locating this bug.
- Crystal Contacts and Unit Cell (in the Tools tab) were not working when using Java, starting in FirstGlance 2.4.
- Clicking an atom when displaying Crystal Contacts or Unit Cell (in the Tools tab) reported the Element in multiple copies.
- Clicking on Be (beryllium) in 4znl, or Se (selenium) in 4hik reported "(Epsilon, 5th)".
- Certain large models were showing red crosses on every water in the initial view (2q08, 3ai7).
- Clicking on atoms with lower case chain identifiers failed to generate a report (1s5l=1S5L).
- In Contacts & Non-Covalent Interactions (Tools tab), some covalent bonds between the target and contacting atoms were not being shown consistently. Example: in 1g9m, the covalent bond between NAG697 and ASN197 was not being shown unless Show hydrophobic interactions was checked.
-
Bug fixed:
- In the Find.. dialog, the report of atoms found failed to be displayed in Version 2.4.
-
Bug fixed:
- Version 2.4 broke the help panel (lower left panel) for Center Atom, Thin Backbone, N->C Rainbow (and perhaps others), showing "undefined".
Version 2.4 provides updates and bug fixes, especially related to ConSurf.
-
Improved
- Clicking on an atom now reports the chemical element in addition to the atom ID. The element was previously reported in the hover message.
- When slab is on, dragging at the right edge of Jmol no longer moves the slab plane. It has been restored to zooming as usual. A new gesture, Alt+Shift+drag up/down, now moves the entire slab towards or away from the viewer.
- The help for the water toggle button now adds "Protein crystals are highly hydrated, ranging from about 30% to 70% water in different crystals." The More.. link there provides literature citations.
- Triple clicking seems to be needed for distance/angle measurements (only when using Java). The Tools tab Distances/Angles help now mentions this.
- The documentation for
one-letter codes for amino acids
is more detailed and a reference is cited.
Updated: - The most popular browsers have been re-tested in Windows 7, XP, and OS X, and
browser compatibility information has been updated. Notably, Google Chrome
no longer supports Java, and the new Edge browser in Windows 10 does not support Java.
Also, a change in Chrome 47, December 2015, caused JSmol to be unable to rotate molecules until
Bob Hanson made a compensating change in JSmol. Finally, the Maxthon browser has been included.
All tested browsers can now be spoofed. Spoofing simply causes FirstGlance to show the relevant
recommendations -- it does not cause the browser being used to emulate another browser.
- Spoof Chrome*
- Spoof Edge
- Spoof Firefox
- Spoof Internet Explorer
- Spoof Maxthon (JSmol recommendations are more urgent in Windows)*
- Spoof Opera*
- Spoof Safari
* Recommendations are more urgent when the molecule has >10,000 atoms, e.g. 7ahl. -
The visitor counter had been courtesy Sitemeter.Com since February 8, 2006,
recording 583,670 visitors (approximately 1.3 million page views).
Due to problems with Sitemeter, the counter was changed to
StatCounter.Com, setting the initial count to the final Sitemeter count. This change was
made online in the previously released FirstGlance 2.32 on November 12, 2015.
ConSurf related Bugs fixed: - The Backbone, Cartoon and Spacefill buttons in the ConSurf control panel did not restore ConSurf colors after they were changed in the FirstGlance control panel.
- The Backbone, Cartoon and Spacefill buttons in the ConSurf control panel did not hide water that may have been displayed in the FirstGlance control panel.
- The Backbone and Cartoon buttons in the ConSurf control panel did not turn off all renderings that might have been used in the FirstGlance control panel (e.g. secondary structure rockets or traces).
- The Reset button in the ConSurf control panel did not restore the gray color of backbones of chains not colored by evolutionary conservation.
- The 4 views were displaying unwanted water due to remediation in the Protein Data Bank which assigned chain names to water.
- ConSurf was reporting sequence identity between some non-identical chains (e.g. chains A and P in 2vaa).
- When Sequence Numbers or Residue Names were checked, in most Views, they were colored by chain rather than by the color scheme of the View (e.g. N->C Rainbow).
- The ConSurf Colors checkbox was inappropriately greyed out for some Views (e.g. Thin Backbone).
- The ConSurf Colors checkbox was inappropriately greyed out when Sequence Numbers or Residue Names checkboxes were used.
- The Isolate tool in the FirstGlance control panel was turning off ConSurf coloring.
- Dots in view 3 remained yellow after view 2.
- Toggling the Spin, Background or Quality buttons in the ConSurf control panel did not propagate those button states to the Molecule Information Tab or the Tools Tab in the FirstGlance Control Panel.
- Dots were not being shown around the balls and sticks of ligands.
Other Bugs fixed: - With certain large molecules (for example, 7ahl), in JSmol, Firefox would freeze for about one minute when the first operation was requested in FirstGlance (for example, toggle spinning off). Thanks to Bob Hanson for fixing this bug in JSmol!
- Center visible moieties in the Hide.. dialog was broken in FirstGlance version 2.2.
-
Updated:
- Browser compatibility and performance with Jmol has been updated, and now includes Microsoft Edge, a browser first released in Windows 10.
- Browser advice: Advice on which browser to use
has been updated to handle the Edge browser.
Recommendations appear in yellow
banners, and can be seen, without installing multiple browsers,
by spoofing a particular browser:
Bugs fixed: - The certificate for the Jmol Java applet was inadvertantly revoked on October 15, 2015, causing operation of all earlier versions to be denied.
- In JSmol (HTML5, no Java) the JSmol menu could not be opened by clicking on the frank or right-clicking in JSmol. This longstanding bug has been fixed thanks to Bob Hanson.
- In the help for Sequences, the links to Uniprot sequences stopped going directly to the sequences section (due to a change at Uniprot).
- The Google Chrome browser stopped supporting Java on September 1, 2015. This release of FirstGlance intercepts a request to use Java in Chrome, suggesting that you use a different browser (Internet Explorer, Firefox and Safari all support Java), or use FirstGlance without Java.
-
New:
- Sequence numbers and residue names can now be displayed for all residues. New checkboxes are located at the bottoms of the Molecule Information Tab, Views Tab, and Tools Tab, just below the Labels (S-, X, ?) checkboxes.
- An index to all the capabilities of FirstGlance, and where to find them, has been created: Where? How? is linked at the bottom of every tab except Preferences, and underneath the molecule.
- A new Practical Guide to Homology Modeling, which also tells how to find X-ray or NMR
models, is now linked to the first page at FirstGlance.Jmol.Org, and in the Resources tab.
Bugs fixed: - Putting halos on some atoms (with Find..) caused the automatic centering to fail when using Isolate...
- Under Sequences (Molecule Information Tab) the link to PDBe (Europe) has been removed. The PDBe website was restructured and the interactive sequence display seems no longer to be available.
- Reset was not resetting the Labels checkboxes (bottom of every tab except Preferences).
- Crystal Contacts (Tools tab) was not showing any balls for contacting nucleotides that lacked a phosphorus atom (5' terminal nucleotides). The c5' atoms of those nucleotides are now shown as balls. For example, 14 nucleotides contact the asymmetric unit of 1d66. 12 of those have phosphorus atoms; 2 lack phosphorus atoms. Prior to the bug fix, only 12 nucleotide balls were shown.
-
New:
- A new dialog, Isolate, will be found between the previously available Hide and Find (near the bottoms of the first three tabs). Isolate makes it easy to hide everything except one entity. Isolate can be used in combination with Hide, in either order.
 Molecular animations are
demonstrated in Powerpoint®.
These animations for Powerpoint were generated with the PolyView-3D Server
recommended in
Presenting Views from FirstGlance, which can be
accessed from a link at the bottom of the lower left panel in FirstGlance.
Molecular animations are
demonstrated in Powerpoint®.
These animations for Powerpoint were generated with the PolyView-3D Server
recommended in
Presenting Views from FirstGlance, which can be
accessed from a link at the bottom of the lower left panel in FirstGlance.
Improved:- Hide can now hide atoms with halos (namely, atoms that were given halos with Find). This is useful when multiple copies of the same thing need to be hidden. For example, 1or9 is an 18-chain assembly. (Use Java for this large structure.) Each chain has a helical domain and a globular domain. To hide all 18 globular domains, Find sequence range "61-199". Then Hide atoms with halos.
- Hide now informs you when you try to hide something that is not present in the model. (Previously, it said nothing while the molecular view remained unchanged.)
- The term chain is now linked to an explanation.
- An explanation of backbone traces was created in Proteopedia, and linked wherever backbone traces or ribbons are mentioned.
- In the Chain Details help (Molecule Information Tab), individual chains can now be highlighted by clicking on the chain name.
- The Vines view now remembers its checkboxes until the end of a session.
- For biological unit models from the MakeMultimer server, the help was improved for Biological Unit under the Resources tab.
- For NMR models, the help was improved for Local Uncertainty under the Views tab.
- The "smaller ligands+" checkbox in the ligands+ button help is now remembered for the duration of a session.
- A change in Jmol long ago has been causing the initial zoom to be too high when Jmol is rectangular. This has been fixed (set zoomLarge false).
- Alternate locations are now discussed in the help for the Vines view, where they can best be seen.
- The Gallery has some new cases (1ijw, 3hyd and 3if7).
- The text in the Snapshot Gallery was updated. The snapshots themselves have not yet been updated.
Bugs fixed: - Centering of the visible atoms often did not work. (Thanks to Bob Hanson for fixing this bug in Jmol.)
- The tool Salt Bridges / Cation-Pi Interactions failed to warn that some interactions are not shown when relevant amino acids are missing, or have incomplete sidechains. The new warning has a yellow background.
- The Crystal Contacts and Unit Cell tools now behave appropriately for models that lack crystallographic information, including homology models from Swiss-Model, biological units from MakeMultimer, and models from the Orientations of Proteins in Membranes server.
- Homology models from Swiss-Model, when uploaded, generated a large spurious alert message.
- Homology models from Swiss-Model, when uploaded, had a garbled report in the molecule information tab.
- Some models from the Orientations of Proteins in Membranes server (available in the Resources Tab) had garbled reports in the molecule information tab, and also produced a large spurious alert during loading. Example: 4fz0.
- Models from the Orientations of Proteins in Membranes server (available in the Resources Tab) contain pseudo-atoms marking the predicted boundaries of the hydrophobic portion of the lipid bilayer. In the View: Vines, with more detail checked, when the ligands were hidden with the Ligands+ button, pseudo-atoms close to protein remained visible as erroneously covalently bonded to protein.
- Hide, Atom: when the atom was calcium (e.g. 4aqr) some results were incorrect.
- Reset was resetting most things even when canceled.
- Reset was not obeying the spin preference.
- Reset was not turning the Quality button off.
- The "smaller water" checkbox in the water button help was being remembered but not being obeyed.
-
New:
- Preferences:
- Spinning when the session begins is now optional.
- When using JSmol, you can hide the suggestions for browsers
that perform better. Exceptions:
- Because the performance of Internet Explorer with JSmol is unacceptably slow, the suggestion not to use it cannot be hidden.
- Because rotation in Opera is unacceptably slow/jerky, the suggestion not to use it cannot be hidden.
- When using JSmol, you can hide the recommendation to use Java for better performance.
Improved: - Help and explanations have been clarified in several places,
especially for Crystal Contacts (Tools tab).
Bugs fixed: - Atoms that were hidden were nevertheless reported when touched with the mouse (onMouseOver), or clicked. This caused mis-identification of atoms, and selection or hiding of incorrect moieties with mouse clicks. (Jmol bug, thanks to Bob Hanson for the fix.)
- Water hydrogen atoms were inappropriately displayed when present in the model (1lcd, 1lfa). (Jmol bug, thanks to Bob Hanson for the fix.)
- The Contacts & Non-Covalent Interactions Tool was not working when the PDB file lacked standard header information. Thus it was not working with uploaded PDB files such as homology models, biological units from MakeMultimer, etc. Thanks to Eugenia Polverini (University of Parma, Italy) for calling this bug to my attention.
- In the Contacts & Non-Covalent Interactions Tool, selecting a sequence range starting with the higher sequence number and finishing with the lower sequence number did not work.
- The number of missing residues reported was incorrect when there are multiple models (e.g. 2q3o). The sum for all models was reported rather than the number missing in the first model.
- The "Get Biological Unit" buttons were going to a submission form instead of the final result for the current PDB code, due to a change at the MakeMultimer server.
- PDB files lacking SOURCE records caused FirstGlance to fail to load.
- Clicking on a water oxygen atom incorrectly identified it as "(main chain)". This has been fixed for both HOH and DOD (deuterium oxide).
- The amino acids graphic chart was not displayed in the section on amino acids in the Notes.

Version 2.00 mostly concerns making the use of
JSmol (no Java, the new default) more user friendly.
-
New:
- Default: no Java. This version defaults to using no Java (using JSmol and HTML5). Because JSmol (no Java) is significantly slower than Jmol (Java), a "BUSY" message appears over the molecule while commands are processed. Jmol (Java) remains an option in Preferences, or with a checkbox at FirstGlance.Jmol.Org.
- Browser advice: The Chrome browser is recommended when using JSmol because it has the best performance. Internet Explorer (extremely slow overall) and Opera (very slow rotation) are discouraged. Recommendations appear in yellow banners, and can be seen, without installing multiple browsers, by spoofing a particular browser:
- Preference: A new tab Preferences has a checkbox to set using Java (Jmol_S, the signed Jmol applet) as the default. Query parameters &JAVA or &NOJAVA override this preference. The checkbox at FirstGlance.Jmol.Org uses query parameters.
- Large molecules: When the model contains 10,000 atoms or more, using Java (Jmol_S, the signed Jmol applet) is recommended for better performance.
- Multiple models: When using JSmol (no Java), and when the data file contains multiple models (typically for NMR experiments), only the first model is loaded. This avoids overloading JSmol. Currently there is no "View All Models" capability when using JSmol, but this is planned for a future version of FirstGlance. When using the Jmol_S signed Java applet, all models are loaded, and "View All Models" is available.
-
Installing and Enabling Java
has been explained in detail for Windows and OS X and all
popular browsers.
Bugs fixed: - The Labels Front checkbox was not working.
- Some links to Troubleshooting were broken.
Version 1.98 is a major upgrade: the Jmol Java applet
is now always signed. Optionally, FirstGlance will run
without Java using JSmol.
-
New:
- Signed Java Applet. This version defaults to the signed Jmol applet. This was done because Oracle has announced that unsigned Java applets will soon be prohibited for security reasons.
- Without Java.
This version can also operate with
JSmol,
instead of the Jmol Java applet.
JSmol does not require Java, which has become increasingly
problematic. (The JS in JSmol stands for javascript.)
Performance (smoothness of rotation, speed of responses)
is somewhat diminished in JSmol, but adequate except for the largest
structures (thousands of atoms).
Bugs fixed: - "Center visible chains" was not always working.
- Some hydrogen atoms, and one nitrogen in histidine, were not being colored in the Charge view. (Protein amino terminal charges are still not shown due to difficulty in identifying them.)
- When a standard residue occurs as a ligand, the count reported
in the Molecule Information Tab included occurrences in protein
or nucleic acid, not just ligand (e.g GLY in 3f4j, 4cpa; A in 1bkx).
OCA: - Uses of OCA removed in version 1.95 have been restored.
- The signed Jmol applet is used for Mac OS 10.6 (Snow Leopard), Mac OS 10.5 (Leopard), and Mac OS 10.4 (Tiger). This works around what appears to be a bug in Java version 1.6.0_51. This change does not affect Windows or Mac OS 10.7 or later, which use Jave 1.7.
-
New:
- Two new Tools, Crystal Contacts and Unit Cell, have been added. Further options for these views are planned, but the basic views appear to be working.
- A new View, Solid, has been added. It is the same as the initial view in the tool Contacts & Non-Covalent Interactions, but with a new explanation and examples.
- The molecule information tab now has a line about the presence
or absence of hydrogen atoms in the model. It is linked to a new
help panel with details, and an introduction to hydrogen atoms in PDB files.
Enhanced: - Hydrophobic/Polar (Views tab) was improved. Cys is now deemed hydrophobic. Most importantly, non-standard amino acids are colored white (formerly magenta).
- Local Uncertainty (Views tab) now gives the minimum, average, and maximum temperature for each chain, water, ligands+, and hydrogen. The chain with the lowest average temperature among a group of sequence-identical chains is highlighted in boldface.
- Slab now has a 20% thickness option, as well as the previously available 10%, 6%, and 3%. Also there is a new checkbox which unhides the back of the molecule, so you can see the inside of a cavity or channel.
- The report by Disulfides/S/Se is more detailed and includes a count of sulfur in non-standard residues. There are also many more examples listed in the help.
- Instructions for obtaining Biological Units now offer a backbone
only model, useful when the all-atom model exceeds Java memory.
Bugs fixed: - Some buttons (such as Spin and Background) were sometimes inappropriately canceling centering, hiding, or target selection in Contacts.
- Non-standard amino acids were being colored "polar" (magenta) in the Hydrophobic/Polar view. They are now white.
- Some selenocysteines were not being counted by Disulfides/S/Se.
- Selenium and iron (Se, Fe), when clicked, were being inappropriately described as "Epsilon, 5th".
- OCA has been out of service for weeks, so some links that use OCA have been removed or directed to other services. Removed: FASTA sequences, OCA under Resources. Redirected: View PDB file now fetches from RCSB.
-
Additions:
- The Hide.. and Find.. tools are now available in all four tabs (at the bottom). Thanks to Frieda Reichsman for this suggestion.
- Secondary Structure (Views tab) now reports the percentages of helix, strand, and neither.
- Charge (Views tab) now reports the numbers of positively and negatively charged amino acids with incomplete sidechains, in addition to the total numbers. The counts now exclude hidden residues and alternate locations and include the carboxy termini. Previously carboxy termini were overlooked in the Charge view, but now they are colored red for negative charge. (Amino termini are more complicated and will be added in a later version.)
- Atoms with alternate locations, when they occur, are
now reported in the Molecule Information Tab, with a tool
to find them. Examples: 3hyd, 1bsz.
Changes: - The initial background is black instead of white, to make the "empty baskets" for missing residues more visible.
- The Spin, Background, etc. buttons have been rearranged to make them
more consistent between tabs.
Bugs fixed: - Hide and Find were missing from the molecule tab when the molecule lacks a proper PDB file header (such as PDB files from MakeMultimer or Orientations of Proteins in Membranes).
Version 1.9 is a major upgrade of FirstGlance in Jmol.
- Controls have been reorganized into 4 sections, each with its own tab: Molecule, Views, Tools, and Resources.
 The initial view now marks regions with missing residues
with an "empty basket" (see image at right). The empty baskets
can be hidden with a checkbox in the Missing Residues report,
available from a link the Molecule Tab.
The initial view now marks regions with missing residues
with an "empty basket" (see image at right). The empty baskets
can be hidden with a checkbox in the Missing Residues report,
available from a link the Molecule Tab.
- The molecule tab (titled with the PDB code when applicable) offers
information from the "header" of the PDB file:
- Name of the molecule.
- Dates of deposition and release.
- Method of structure determination.
- Resolution, R value and free R. Resolution and free R are graded by FirstGlance to help novices interpret these values.
- Asymmetric unit:
- The number of chains, and the number of sequence-distinct chains. Clickable chain names identify the location of each chain.
- Details about each chain including its length, whether protein, DNA or RNA, presence of non-standard residues.
- Sequences are easily accessible from links to various sequence databases. (There is still no clickable sequence-to-structure listing -- it remains under development.)
- The total number of missing residues. Also a summary table listing the residues missing in each chain, and their sequence numbers, and the number of charged residues that are missing. (The S2C "gaps" server is no longer necessary.)
- The biological unit is readily accessible.
- Ligands and non-standard residues are listed with their counts and full names. Clicking on one shows its position(s) in the model.
- The publication abstract at PubMed is one click away.
- All literature citations given in the PDB file are listed, with clickable PubMed ID and DOI links.
- The PDB file text is one click away.
- The Views Tab offers display and color schemes that affect the entire structure. It includes the 8 views previously available, plus Local Uncertainty (formerly under More Views) with expanded help and options.
- The Tools Tab offers tools that identify, highlight, or measure specific substructures: Disulfides/S/Se, Salt Bridges/Cation-Pi, Contacts, Distances/Angles, and Advanced/Technical. None of these are completely new but there are some new options and expanded explanations.
- The Resources Tab offers guidance for use of servers external to FirstGlance, some of which were formerly available under "Key Resources". Guidance has been greatly expanded. In addition to the previously featured resources (biological unit, evolutionary conservation, and Proteopedia), there is now guidance for all-atom clash analysis with MolProbity, prediction of intrinsically unstructured regions of protein chains, and marking of lipid bilayer boundaries for integral membrane proteins.
- Hide.. and Find.. are now available at the bottoms of any
of the four tabbed control panels. There are some new capabilities in these
tools:
- The Find.. tool now finds amino acid or nucleotide sequence fragments, e.g. sequence=skek or sequence=cgg.
- The Hide.. tool now asks whether you wish to hide all copies of the residue/group or atom you clicked, or only the single one clicked.
- Formerly, FirstGlance showed structures but did not report counts. This version, for the first time, reports counts, e.g. number of chains, amino acids and nucleotides, missing residues, ligand groups, non-standard residues, disulfide bonds, sulfur atoms, selenium atoms (selenomethoinine and selenocysteine), positive and negative charges, and numbers of atoms/residues found with the Find.. tool.
- Bugs fixed:
- In the Hide.. tool, with the range option, formerly when the higher sequence number was clicked first, hiding failed.
- In the Hide.. tool, with the Atom option, the red list of atoms hidden inadvertantly included the XYZ coordinates of the atom (evidently due to a change in Jmol). Also clicking the same atom twice (possible even after it is "hidden" in some displays) was not properly handled.
- In the Find.. tool, there were problems when the query included a single quote, as in *.c5'.
- The Backbones checkbox in Contacts Shown was not being remembered.
- Operation in the Internet Explorer 9 web browser has been restored
after failing for several months. (Operation in Internet Explorer 7 and 8,
Firefox and Chrome never failed.)
Operation in IE9 was accomplished with the following restructuring:
- FirstGlance now uses Jmol.js for the first time.
- The division layout (file fg.htm) was restructured following the resizable Jmol scheme kindly provided by Angel Herráez.
- Java security risks and recommendations for using Java safely have been documented.
-
Zoom help was expanded to include moving the cursor to the right inside
edge of Jmol. Mac:
 ; Windows:
; Windows:
 .
.
- A link to Mouse Use was added beneath Jmol, along with a link to Java Security.
- Updated list of compatible browsers and notes on browser peculiarities.
- The cutoff distance for van der Waals interactions in the Contacts display has been reduced from 4.5 Å to 4.0 Å at the recommendation of several crystallographers. See Van der Waals interactions.
- Using Advanced Options (at firstglance.jmol.org, or by clicking New Session within an existing session) you can request the signed Jmol applet. This version of Jmol's signed applet enables writing a .jmol file that contains everything (including the state script and the atomic coordinates) needed to regenerate the view at the time the .jmol file was written. The .jmol file can be dragged into the Jmol application, or the signed applet. To write a .jmol file, click on Jmol at the lower right of the molecule to open Jmol's menu, then open the Console. In the lower white box, issue the command "write filename.jmol".
- Options for visualizing Biological Assemblies have been expanded to include the MakeMultimer Server, and to caution about the frequent apparent errors at the PISA server. (These changes were added to version 1.45 on May 7, 2010.)
- Added a color key to Color by Uncertainty (under More Views..).
- In Contacts.., added a caution that salt bridges and cation-pi interactions are shown only for protein.
- A method for calculating pI and charge at various pH values was documented, and linked into the Charge.. dialog. (This and the previous two changes were added to version 1.45 in May/June, 2011.)
- Added more information under "How Does It Work?" in What Is FirstGlance in Jmol?.
- Noted in Adoptions that Nature discontinued direct 3D View links to FirstGlance as of the September 23, 2010 issue. (This and the previous item were added to version 1.45 in February, 2012.)
- Under More Views.. is a new dialog to show disulfide bonds clearly, as well as to show cysteine residues not participating in disulfide bonds. Also cys residues can be labeled.
- Under Key Resources.. PQS (Probable Quaternary Structure) has been updated to use PISA, as well as offering PQS and RCSB for obtaining biological assemblies.
- Some progress was made in preparation for translation to other languages, but FirstGlance is still not translation-ready.
- Jmol now resizes automatically with the browser window.
- Added Key External Resources, replacing the 4 buttons "PQS, PDB, OCA, Gaps?". New on the list are Proteopedia, Evolutionary Conservation, and MolProbity.
- Changed Secondary Structure. It now represents beta strands and sheets in a more realistic fashion -- gone are the straight planks shown in earlier versions. Also the ribbons are a bit narrower in Cartoon.
- The initial view for Charge.. is now to color only the charged atoms. All the previous options remain available.
- Status HPUB at the PDB, or an invalid PDB code, is detected, and explanatory help should appear automatically. (A similar feature was present in version 1.02, but was later broken by changes at the PDB, and regardless, had not been installed in versions higher than 1.1.)
- Incorporated links to Proteopedia.Org in several places, with explanations of why it is useful in a particular context.
- Limitation removed: Atoms can now be identified with Slab on, due to improvements in Jmol. Identification is by clicking on the atom (which reports at the lower left of Jmol) or by touching the atom, which reports in a yellow "hover" box. Slab mode can also be used in the Contacts and Hide dialogs now.
- Protein atom identifications now include "main chain" or, for sidechain atoms, the greek letter, spelled out in English.
- Included "Atom" in hover report (e.g. "...Element=C, Atom=C7"). It was previously included in the identification report produced by clicking on an atom.
- The extensive Notes document was updated throughout. Some more technical documents have not yet been updated, but are marked as out of date (e.g. those listed under Technical Information for Programmers).
- Bug fixed: thymidines were erroneously labeled as having incomplete sidechains "S-" due to a change in nomenclature with the remediated PDB format (t.c5m is now t.c7).
- Bug fixed: A351 in 1bkx is not covalently bonded to any other nucleotide. It was not displayed within the "ligands+" group as intended.
- Bug fixed (I think): FirstGlance occasionally failed to complete loading. This was found to happen when the server supplying the visitor count (sitemeter.com) failed to respond. The meter code was put inside a division, which appeared to permit loading even without a response.
- Bug in Jmol fixed: in "Center Atom" mode, clicking on Jmol's background no longer causes a zoom out.
- Bug in Jmol fixed: hydrogens on charged sidechain nitrogens are now colored blue in the Charge.. display, "color only charged atoms". Formerly they were white.
- Adoptions was updated to include Protein Science.
- Work was begun to prepare FirstGlance in Jmol for translation to español and other languages.
Limited release for the Epitopia server.
- Added graphics Quality toggle button.
- Added support for the Epitopia Server.
- The Adoptions page was updated to include Proteopedia and TOPSAN.
- Updated help for taking snapshots for presentations.
and version 1.01 (at firstglance.jmol.org, Jmol applet remains 10.2.0).
- Fixed a bug that prevented hiding (in Hide..) or selecting (in Contacts..) chains of DNA. This bug was created when the Protein Data Bank changed the PDB format on August 1, 2007, in accord with their data remediation project. Specifically, the former nucleotide names A,C,G,I,T,U now designate only RNA, while DNA nucleotides are named DA,DC,DG,DI,DT,DU.
- Fixed a bug that prevented the Labels: Front checkbox from working. This had resulted from a change in Jmol. (This fix applied only to version 1.39.)
- Support for the Pepitope Server.
- Support for the Selecton Server.
- *Version 1.36 [April 7, Jmol 11.1.27] and 1.37 [April 13, Jmol 11.1.27] were installed at only the Pepitope and Selecton servers. Version 1.0 (Jmol 10.2.0) remained installed at firstglance.jmol.org, and version 1.2 (mislabeled 0.995, Jmol 10.2.0) remained at ConSurf and OCA.
- * Version 1.38 [August 27, Jmol 11.3.11] was installed for Pepitope, Selecton, and at the OCA server (used for uploading PDB files saved from Pepitope or Selecton jobs). Version 1.0 (Jmol 10.2.0) remained installed at firstglance.jmol.org.
- In Jmol 10.2.0, calcium was incorrectly classified as protein. This bug was fixed in later Jmols, enabling the removal of the workaround in FirstGlance that was needed previously.
- Preliminary support for submitting a Jmol state script to generate high-resolution images at the ConSurf Server only.
- Added Polyview-3D to Presenting Molecular Views. (August 6, 2007)
- Dec. 10: After seven months of service, no major problems came to light in version 0.991, so I deemed it version 1.0 with no further changes. Version 1.0 remains the main public version at this time.
- Jan. 14: Added What Is FirstGlance in Jmol? (Thanks to Phil Leader for suggesting this.)
- Jan 14: Added instructions for copying static snapshots of molecular views and pasting them into presentations or documents. Also mentioned MolSlides in Jmol. The new instructions are linked to the bottom of every help panel.
- Since version 0.991 is now version 1.0, version 0.995 should have been called version 1.2.
- Version 0.995 (1.2) is public, at this time, only at the ConSurf Server and OCA. (The version at OCA was updated to 0.995 to support upload of saved ConSurf job PDB files.)
- This version was released at the ConSurf Server and OCA Servers only. It was not released at firstglance.jmol.org pending further improvements. Release at the OCA Server was to support display of saved and uploaded ConSurf PDB files.
- Added support to display evolutionary conservation results
from the ConSurf Server.
- Support for ConSurf increased the size of FirstGlance by about 25% (from 15K lines in .htm and .js files to 20K). Most of the code was adapted from Protein Explorer. More..
- This involved the first use of messageCallback from Jmol to javascript, which enabled the following change, and will enable future enhancements.
- The "Please Be Patient!" help that appears at the beginning of a session is now automatically replaced with the Introduction after the molecule appears.
- Moved the visitor count from the "Please Be Patient!" help (which now may disappear so quickly the count might fail to load) to beneath the molecular view (scroll down).
- Added a new link to the control panel: New Session.
- The Charge.. default color scheme has been changed to make it easier to see the distribution of charged atoms. The old 5-color scheme remains available via a checkbox.
- One of the new Charge.. views shows only charged atoms. Because some crystallographic models have incomplete sidechains, where the charged atoms are missing, these are detected and indicated.
- Hide.. has a new option to hide a range of residues within one chain (or multiple such instances). This is similar to the range-targeting function previously available in Contacts.
- Hide.. has a new Invert Hiding checkbox. Previously hidden moieties are displayed, and vice versa. This makes it easier, for example, to view a single chain out of many (first hide it, then invert hiding).
- Added instructions for copying static snapshots of molecular views and pasting them into presentations or documents. The new instructions are linked to the bottom of every help panel.
- Bug fixed: In Hide.., the option to Center Visible Chain was not working reliably.
- Bug fixed: Following a Reset, certain operations (such as Center Visible Chains in Hide..) had inappropriate consequences (such as an inappropriate start of spinning, or failure to center properly).
- Bug fixed: After showing Contacts, and then using Center Atom.., clicking Return to Contacts failed to clear centering mode.
- Bug fixed: Jmol defaults to showing hydrogen bonds for rare PDB files. These are now hidden with an explicit "hbonds off". An example is 4MDH (in FGiJ 0.991). See Jmol Bugs.
- Clicking the "Zoom Up" button no longer intereferes with target selection for Contacts, and does not interfere with hiding a range of residues, by displaying its help.
- Added a color key for the element-colored displays of salt bridges and cation-pi interactions (under More Views..).
- Added the journal Molecular Biosciences to the Adoptions page.
- Non-standard residues are now shown as crosses ("stars" in Jmol command language) by default. The crosses can be hidden in More Views.., or Ligands+. This change was made in accord with the goal of not leaving important information invisible. Crosses were chosen because they don't distract too much from the backbone displays when there are many non-standard residues, as for 1EVV. Examples: 1EVV, 1BKX.
- New Exit buttons were provided for Hide.. and Center Atom.., replacing the former exit links.
- At the main page, enabled pressing Enter (in addition to clicking the Submit button) to submit a PDB code to start a FirstGlance session.
- Fixed a bug that prevented the atom identification report from displaying, to the lower left of the molecule, when the first operation performed in the session was clicking on the molecule.
- Fixed a bug in atom identification reporting that occurred only in Mac Safari with the latest java update. This bug caused the Safari window to disappear suddenly, at least after a recent java update was released by Apple.
- Fixed several bugs that prevented the PDB identification code from showing, to the upper left of the molecule, after the background color changed.
- Minor improvements/corrections in documentation.
- Contacts.. is a new option to show atoms contacting any "target" moiety, which is selected by clicking on the molecule, or with the new Find.. option (see next item). Four views of atoms contacting the target are available, with increasing levels of detail. Distances can be measured between putatively non-covalently bonded atoms. Hydrogen atoms (if present in the model) can be displayed. Seven kinds of non-covalent bonds can be displayed or hidden independently of each other.
- Find.. is a new option that puts yellow halos around atoms designated by sequence number, 3-letter residue abbreviation, or element name. The halos are visible in all views.
- A new Snapshot Gallery was provided.
- More Views.. has a new option to show all salt bridges and cation-pi interactions throughout the molecule.
- Isolated standard amino acids or nucleotides, invisible in the "backbone" displays of previous versions, are now spacefilled in the initial view, and included in Ligands+ (More..). Selection of these was made possible by a new Jmol command, "connected" (More..).
- Solutions were implemented to handle hiding or selecting the single unnamed chain in some PDB files, as well as ligands that have chain designations (such as heme in 2HHD). These are important in the new Contacts.. target selection interface, and they improve the function of Hide.. over the previous release.
- Hide Hydrogen was added to the Hide.. interface. This enables uncluttering ligands or non-standard residues. (Example: 1AL4 has hydrogen on both.)
- There is a new Reset link that attempts to reset the session to the initial view, and most of the initial settings. I've undoubtedly overlooked some things that need resetting.
- Gaps in the coordinates, compared to the sequences crystallized, can be located using new documentation, which links to the "SEQRES to Coordinates" alignment (S2C) resource by Wang and Dunbrack.
- A
 button for accessing the sequences and gaps help, and the link
to S2C (see previous item) was added to the main control panel, next to
the PQS, PDB, and OCA buttons.
button for accessing the sequences and gaps help, and the link
to S2C (see previous item) was added to the main control panel, next to
the PQS, PDB, and OCA buttons.
- Most links and buttons now display pop-up help when the mouse is held over them for a few seconds.
- The inability of Jmol to report atom identifications when slab is on has been handled better. Slab is forced off when selection by clicking is needed (Contacts.., Hide..). When Slab is on, the hover and echo identification reports are replaced with a "Sorry..." message.
- Slab thickness can now be controlled (3 steps).
- The definition of "carbohydrate" was greatly improved (for the "hide carbohydrate" checkbox). See
- Rare Anomalous atoms are no longer invisible, but are shown as dots.
- Hidden atoms are now listed by element, sequence number, and chain, rather than by serial number. (Serial number is used internally.)
- Specifications for a future interactive sequence listing were developed.
- Updated many help documents, including and Known Unresolved Issues. Added a section on selection of unnamed chains to Technical Information. A new list of Jmol Bugs and Limitations gathers together information scattered in other FirstGlance documents, and buried in its javascript/Jmol script code.
- The help for N->C Rainbow now includes a spectral color sequence.
- Added a "mascot" image to the main page.
- mmCIF format atomic coordinate files can now be loaded instead of PDB format files, using a new Advanced Option available from the main page. CIF files do not work properly in FirstGlance. CIF loading is provided for testing purposes only. See Preliminary CIF Observations.
- Bug fixed: The axes, unit cell, and boundbox checkboxes under More Views.. did not work in Gecko browsers in FGiJ 0.98.
- Thanks to PDB files can now be uploaded for display in FirstGlance in Jmol. There is a new option on the main page to do this. for inserting such an upload slot into any web page are provided.
- Added a new view option Hide.. that hides clicked chains, residues/groups, or atoms; or all protein, DNA, RNA, or carbohydrate. The hidden moieties remain hidden in all views, until explicitly re-displayed.
- Added a PQS "button" in the control panel (upper left panel, near the PDB and OCA buttons).
- More Views.. now includes a new section on evaluating model quality, with "color by temperature" (thanks to Gale Rhodes) and a link to MolProbity.
- More Views.. now includes checkboxes to show the axes, unit cell, or bound box.
- More Views.. now includes a link for visualizing dipole moments in Jmol.
- Added 1IGT (antibody) to the Gallery to provide an example with disulfides in multiple chains, and both inter- and intra-chain disulfides.
- Fixed a bug in Vines.., Hide Sidechains that failed to hide the sidechains in nucleic acids.
- Korea was added to the list of mirror sites.
- The advanced option to use browser frames was discontinued.
- The PDB code (or filename) is now the first word in the title of the FirstGlance window. This makes it easy to distinguish windows showing molecules from each other, and from the FirstGlance main page window.
- It is now possible to enter a molecule's URL into a form slot on the main page, to display a molecule from any server (in addition to a PDB identification code).
- Ligands+ are now shown by default in the initial view. The definition of Ligands+ has been changed to exclude most non-standard amino acids and nucleotides. When the Ligands+ button is depressed, or in More Views.. (new, see below), the accompanying help now includes a checkbox to show non-standard residues. The distinction between Ligands+ and Non-standard residues is sometimes incorrect. Notes were added to explain these definitions, and the examples of the problems were documented.
- Disulfide bonds are shown by default in the initial view, in all backbone trace views, and in Vines.
- New entries in the Gallery illustrate disulfide bonds and non-standard residues.
- Backbones are no longer interrupted by non-standard amino acids and nucleotides. (Formerly, these were displayed as Ligands+, which interrupted the backbones.) For more information, see the new notes on non-standard residues.
- In the Vines.. display,
there are now (in the help panel) new checkboxes
to
- Hide sidechains.
- Display more detail: all non-water atoms as sticks,
- Hide hydrogen.
- In the Ligands+ display, there is now (in the help panel) a new checkbox to display smaller ligands+ atoms.
- When water is displayed, there is now (in the help panel) a new checkbox to display smaller water atoms.
- At the time of this release, mouse hover atom identification reports were not working in OSX 10.3.9 (but worked in 10.4 and Windows). Therefore, clicking an atom now identifies it at the lower left of Jmol, and in the browser status line. The report to Jmol required implementing pickCallback(), which may not work in some browsers. A problem remains reporting the element in these reports. The hover report remains, when it works.
- Using clicking for atom identification required changes in
the centering mechanism. The former Center All link
was changed to Center Atom. The help accompanying the latter
offers a link to Center All.
- Turning centering off after a single centering click was not possible, because in "set picking center" mode, Jmol 10.00.46 does not call pickCallback(). Using "center atomno=" instead of "set picking" could have avoided this, but it caused unwanted changes in zoom.
- The present implementation was complicated by the need to turn centering off automatically, when some view links or buttons are pressed by the user before centering was turned off. This was implemented.
- The PDB code shown in Jmol at the upper left now changes automatically to a contrasting color when the background is changed. (Previously, it remained black, becoming invisible on a black background.)
- Known unresolved issues was revised, and unimplemented plans for future enhancements were added.
- Several mirror sites are now listed.
- A new link More Views.. has been added to the upper left control panel. The links to additional viewing options that were formerly in the introductory help have been moved to this new help section, and expanded. New here is information about measuring distances and angles, or opening the Jmol console for entering script commands. In FirstGlance, default units for reporting distances are Ångstroms (Jmol's default is nanometers, rarely used by protein structure scientists).
- The link to Protein Explorer, formerly in the upper left control panel, has been moved to the More Views.. help section.
- The term Structure Explorer is no longer in use at pdb.org. The link formerly of that name has been replaced with a PDB graphic.
- Adjacent to the PDB link, a graphic link to the OCA Browser Atlas entry has been added.
- Fixed bug that reversed the Charge cationic/anionic colors in the color key. (These colors were never reversed in the molecular view.)
- Fixed bug that reversed the Solvent and Ligand colors in the Composition view. (Thanks to Frieda Reichsman for pointing out this bug.)
- Tested dozens of potentially problematic PDB files, documenting the results. See links at the bottom of About FirstGlance.
- Calcium ions are erroneously deemed "protein" by Jmol (through 10.00.48). Other metals appear to be correctly deemed "hetero" and "ligand" (including at least Fe, K, Mg, Mn, Mo, Na, Zn, Cd). The command scripts of FirstGlance were modified for the special case of calcium, to make it display under "Ligands+". (Implemented Feb 4, 2006).
- Tried to prevent problematic atoms from remaining invisible (implemented
February 4, 2006).
- Rarely, Jmol fails to deem standard amino acids as protein. Therefore, "protein" was broadened to include the 20 standard amino acids plus atoms deemed "protein" by Jmol, which (usefully) includes hetero atoms for non-standard residues. Examples. Although the backbone trace displays currently (Jmol 10.00.48) fail to connect the alpha carbons of these amino acids, they are shown spacefilled to avoid leaving these parts of chains invisible. An example is 1AL4.
- Rarely, Jmol fails to assign covalent bonds that should be present. Such groups of atoms are invisible in wireframe (stick) rendering, the normal rendering for the Vines display, which is supposed to show all non-water atoms. Therefore, in Vines, all non-water atoms are shown spacefilled as well as wireframe (or backbone). The diameter of the spacefill is the same as the wireframe (or backbone); hence, when bonded, the spacefilled atoms are hidden within sticks (or backbone). When not bonded, they are visible as small spheres. Examples with many inappropriately non-bonded atoms are 1AL4 and 2SOC.
- Added additional information at the bottom of Technical Information for Developers.
- Added a free visit counter, kindly hosted by sitemeter.com. Clicking on the count (below "Please be patient" when the application starts) reveals referring sites, a world map showing locations of visitors, and browsers and platforms used by visitors, among other useful information.